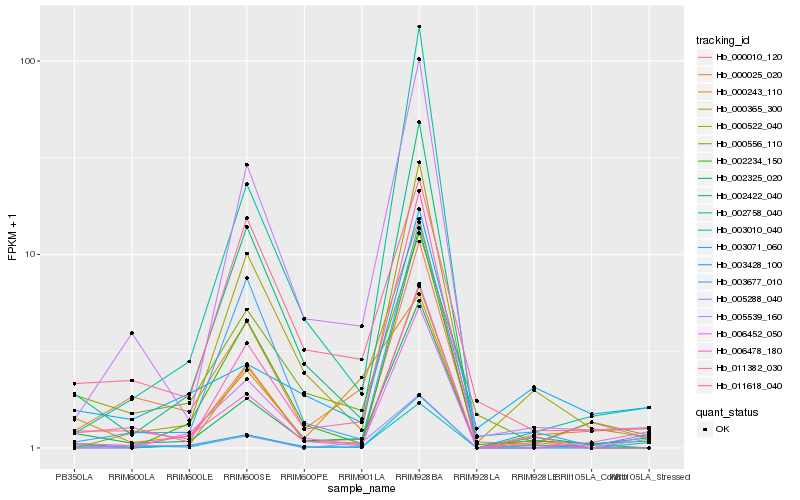
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000025_020 |
0.0 |
- |
- |
PREDICTED: cytochrome P450 71B34-like [Jatropha curcas] |
| 2 |
Hb_002325_020 |
0.1546023813 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 3 |
Hb_005539_160 |
0.1601458147 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 2 member B7, mitochondrial-like [Jatropha curcas] |
| 4 |
Hb_000010_120 |
0.1756055677 |
- |
- |
PREDICTED: cytochrome P450 71A26-like [Jatropha curcas] |
| 5 |
Hb_000556_110 |
0.1907648505 |
- |
- |
hypothetical protein POPTR_0014s06790g [Populus trichocarpa] |
| 6 |
Hb_003071_060 |
0.20152282 |
- |
- |
PREDICTED: beta-galactosidase 15-like [Jatropha curcas] |
| 7 |
Hb_006478_180 |
0.2054055527 |
- |
- |
Nonspecific lipid-transfer protein precursor, putative [Ricinus communis] |
| 8 |
Hb_011382_030 |
0.2106170056 |
- |
- |
Dehydration-responsive protein RD22 precursor, putative [Ricinus communis] |
| 9 |
Hb_002234_150 |
0.2143661746 |
- |
- |
(S)-N-methylcoclaurine 3'-hydroxylase isozyme, putative [Ricinus communis] |
| 10 |
Hb_011618_040 |
0.2156477543 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002422_040 |
0.2163115237 |
- |
- |
hypothetical protein POPTR_0016s02350g [Populus trichocarpa] |
| 12 |
Hb_002758_040 |
0.2164885766 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 13 |
Hb_000522_040 |
0.2175514959 |
- |
- |
Desacetoxyvindoline 4-hydroxylase, putative [Ricinus communis] |
| 14 |
Hb_005288_040 |
0.2183050737 |
- |
- |
DUF26 domain-containing protein 2 precursor, putative [Ricinus communis] |
| 15 |
Hb_003677_010 |
0.2203011492 |
transcription factor |
TF Family: SRS |
PREDICTED: protein SHI RELATED SEQUENCE 1 [Jatropha curcas] |
| 16 |
Hb_003428_100 |
0.2241932099 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 17 |
Hb_006452_050 |
0.2243728585 |
- |
- |
PREDICTED: GABA transporter 1-like [Jatropha curcas] |
| 18 |
Hb_000243_110 |
0.2259567199 |
- |
- |
PREDICTED: WAT1-related protein At3g53210-like [Jatropha curcas] |
| 19 |
Hb_003010_040 |
0.2266233578 |
- |
- |
hypothetical protein AALP_AA7G248200 [Arabis alpina] |
| 20 |
Hb_000365_300 |
0.231319265 |
- |
- |
PREDICTED: cytochrome P450 714C2-like [Jatropha curcas] |