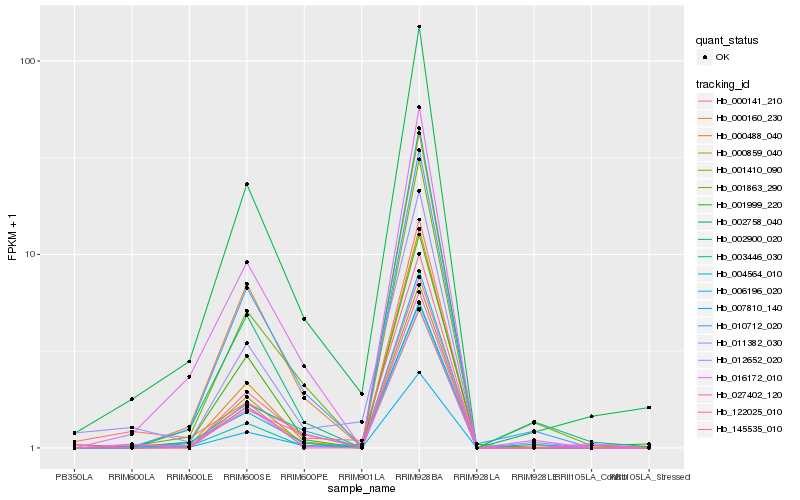
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011382_030 |
0.0 |
- |
- |
Dehydration-responsive protein RD22 precursor, putative [Ricinus communis] |
| 2 |
Hb_002758_040 |
0.1087602344 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 3 |
Hb_145535_010 |
0.1177548859 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |
| 4 |
Hb_003446_030 |
0.1256853417 |
- |
- |
hypothetical protein JCGZ_20198 [Jatropha curcas] |
| 5 |
Hb_027402_120 |
0.1272039801 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 6 |
Hb_000141_210 |
0.1386479621 |
- |
- |
PREDICTED: 36.4 kDa proline-rich protein-like [Camelina sativa] |
| 7 |
Hb_000160_230 |
0.1398284788 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 8 |
Hb_010712_020 |
0.1398376352 |
- |
- |
PREDICTED: caffeic acid 3-O-methyltransferase-like [Jatropha curcas] |
| 9 |
Hb_000859_040 |
0.1416020829 |
- |
- |
Early nodulin 16 precursor, putative [Ricinus communis] |
| 10 |
Hb_007810_140 |
0.1418710811 |
- |
- |
Ankyrin repeat-containing protein [Medicago truncatula] |
| 11 |
Hb_004564_010 |
0.1428041427 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 12 |
Hb_001999_220 |
0.143566461 |
- |
- |
PREDICTED: cytochrome P450 86A1 [Jatropha curcas] |
| 13 |
Hb_000488_040 |
0.1436775274 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 14 |
Hb_002900_020 |
0.1458404525 |
- |
- |
hypothetical protein POPTR_0008s06270g [Populus trichocarpa] |
| 15 |
Hb_016172_010 |
0.146297232 |
- |
- |
hypothetical protein JCGZ_23602 [Jatropha curcas] |
| 16 |
Hb_006196_020 |
0.1467758043 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 100-like [Populus euphratica] |
| 17 |
Hb_001863_290 |
0.1476839213 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Jatropha curcas] |
| 18 |
Hb_012652_020 |
0.1477535197 |
- |
- |
PREDICTED: probable carboxylesterase 18 [Jatropha curcas] |
| 19 |
Hb_001410_090 |
0.1480946176 |
- |
- |
transferase, putative [Ricinus communis] |
| 20 |
Hb_122025_010 |
0.1489377012 |
- |
- |
hypothetical protein POPTR_0003s052201g, partial [Populus trichocarpa] |