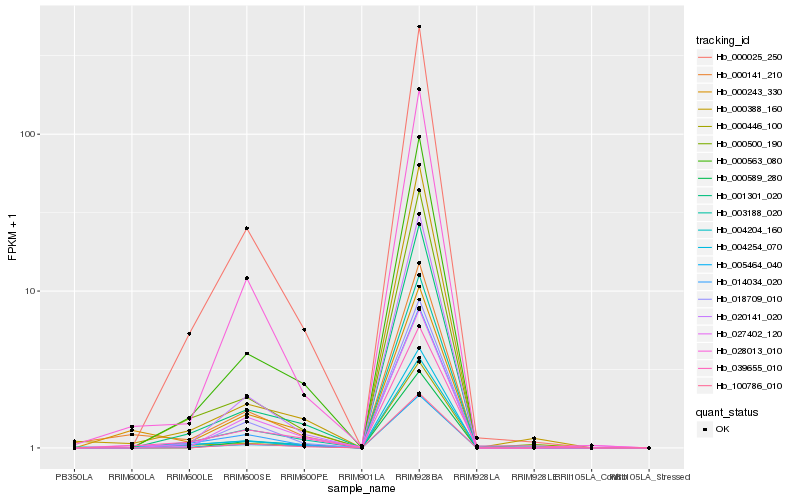
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000141_210 |
0.0 |
- |
- |
PREDICTED: 36.4 kDa proline-rich protein-like [Camelina sativa] |
| 2 |
Hb_000243_330 |
0.0808469128 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_028013_010 |
0.0921218773 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 4 |
Hb_000025_250 |
0.1001256394 |
- |
- |
PREDICTED: flavonoid 3',5'-hydroxylase 2-like [Jatropha curcas] |
| 5 |
Hb_005464_040 |
0.104261771 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 6 |
Hb_000563_080 |
0.1071684585 |
- |
- |
- |
| 7 |
Hb_018709_010 |
0.1071777064 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 8 |
Hb_004254_070 |
0.1075874547 |
transcription factor |
TF Family: NAC |
hypothetical protein POPTR_0015s03700g [Populus trichocarpa] |
| 9 |
Hb_001301_020 |
0.1079817367 |
- |
- |
hypothetical protein JCGZ_06575 [Jatropha curcas] |
| 10 |
Hb_014034_020 |
0.1091772412 |
- |
- |
hypothetical protein POPTR_0075s00200g, partial [Populus trichocarpa] |
| 11 |
Hb_003188_020 |
0.1102532277 |
- |
- |
chitinase, putative [Ricinus communis] |
| 12 |
Hb_020141_020 |
0.1105486388 |
- |
- |
- |
| 13 |
Hb_027402_120 |
0.1112083856 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 14 |
Hb_039655_010 |
0.1114718693 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like [Malus domestica] |
| 15 |
Hb_000446_100 |
0.1127554042 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 16 |
Hb_004204_160 |
0.1159650559 |
- |
- |
S-locus-specific glycoprotein S6 precursor, putative [Ricinus communis] |
| 17 |
Hb_000500_190 |
0.1175990382 |
- |
- |
hypothetical protein JCGZ_21655 [Jatropha curcas] |
| 18 |
Hb_100786_010 |
0.117696513 |
- |
- |
hypothetical protein B456_008G202600 [Gossypium raimondii] |
| 19 |
Hb_000388_160 |
0.1207415081 |
- |
- |
xyloglucan endo-1 family protein [Populus trichocarpa] |
| 20 |
Hb_000589_280 |
0.1220231627 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase 5 [Jatropha curcas] |