| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
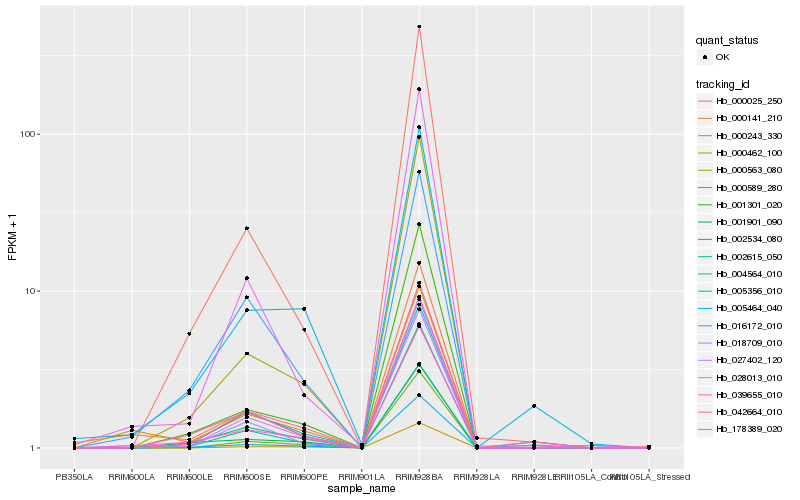
Hb_000243_330 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000141_210 |
0.0808469128 |
- |
- |
PREDICTED: 36.4 kDa proline-rich protein-like [Camelina sativa] |
| 3 |
Hb_027402_120 |
0.1045824073 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 4 |
Hb_039655_010 |
0.1069496806 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like [Malus domestica] |
| 5 |
Hb_005464_040 |
0.122981255 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 6 |
Hb_028013_010 |
0.1289876561 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 7 |
Hb_042664_010 |
0.1306452738 |
- |
- |
hypothetical protein POPTR_0001s07640g, partial [Populus trichocarpa] |
| 8 |
Hb_002534_080 |
0.1308173815 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 9 |
Hb_000025_250 |
0.1327892691 |
- |
- |
PREDICTED: flavonoid 3',5'-hydroxylase 2-like [Jatropha curcas] |
| 10 |
Hb_005356_010 |
0.1337443665 |
- |
- |
hypothetical protein POPTR_0003s16550g [Populus trichocarpa] |
| 11 |
Hb_004564_010 |
0.1348722365 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 12 |
Hb_002615_050 |
0.1362477275 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 13 |
Hb_018709_010 |
0.1362962554 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 14 |
Hb_000589_280 |
0.1366538604 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase 5 [Jatropha curcas] |
| 15 |
Hb_000563_080 |
0.1368720879 |
- |
- |
- |
| 16 |
Hb_178389_020 |
0.1370555956 |
- |
- |
PREDICTED: cytochrome P450 86B1-like [Jatropha curcas] |
| 17 |
Hb_001901_090 |
0.1372368701 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 18 |
Hb_016172_010 |
0.1380307795 |
- |
- |
hypothetical protein JCGZ_23602 [Jatropha curcas] |
| 19 |
Hb_001301_020 |
0.1383918326 |
- |
- |
hypothetical protein JCGZ_06575 [Jatropha curcas] |
| 20 |
Hb_000462_100 |
0.1394118224 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |