| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
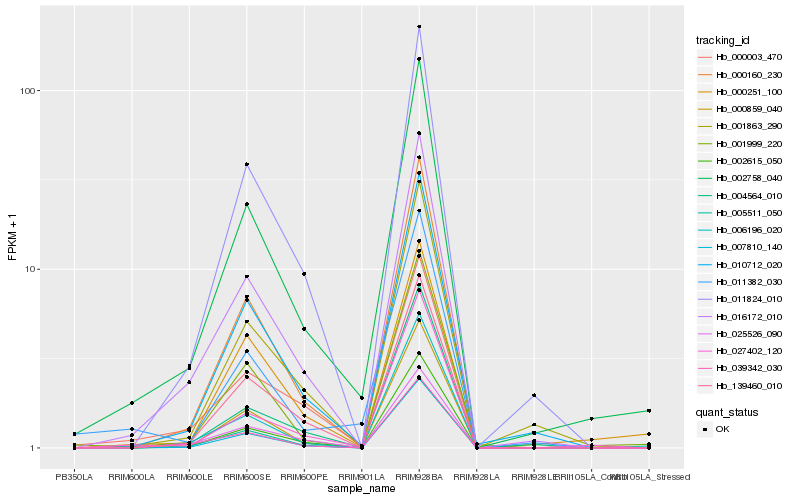
Hb_002758_040 |
0.0 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 2 |
Hb_016172_010 |
0.0864927896 |
- |
- |
hypothetical protein JCGZ_23602 [Jatropha curcas] |
| 3 |
Hb_010712_020 |
0.0930739363 |
- |
- |
PREDICTED: caffeic acid 3-O-methyltransferase-like [Jatropha curcas] |
| 4 |
Hb_000160_230 |
0.0932861224 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 5 |
Hb_001999_220 |
0.0951795152 |
- |
- |
PREDICTED: cytochrome P450 86A1 [Jatropha curcas] |
| 6 |
Hb_027402_120 |
0.0972705623 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 7 |
Hb_007810_140 |
0.0986995092 |
- |
- |
Ankyrin repeat-containing protein [Medicago truncatula] |
| 8 |
Hb_002615_050 |
0.0988727772 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 9 |
Hb_011824_010 |
0.0995337672 |
- |
- |
Metalloendoproteinase 1 precursor, putative [Ricinus communis] |
| 10 |
Hb_005511_050 |
0.1020961678 |
- |
- |
hypothetical protein POPTR_0008s04130g [Populus trichocarpa] |
| 11 |
Hb_001863_290 |
0.1046961933 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Jatropha curcas] |
| 12 |
Hb_000859_040 |
0.1051841766 |
- |
- |
Early nodulin 16 precursor, putative [Ricinus communis] |
| 13 |
Hb_006196_020 |
0.1058140644 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 100-like [Populus euphratica] |
| 14 |
Hb_000003_470 |
0.1077312663 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_039342_030 |
0.1083512066 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_011382_030 |
0.1087602344 |
- |
- |
Dehydration-responsive protein RD22 precursor, putative [Ricinus communis] |
| 17 |
Hb_000251_100 |
0.1088386948 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 18 |
Hb_004564_010 |
0.10933216 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 19 |
Hb_139460_010 |
0.1103994728 |
- |
- |
PREDICTED: beta-glucosidase 18-like [Jatropha curcas] |
| 20 |
Hb_025526_090 |
0.1137350487 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 [Jatropha curcas] |