| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
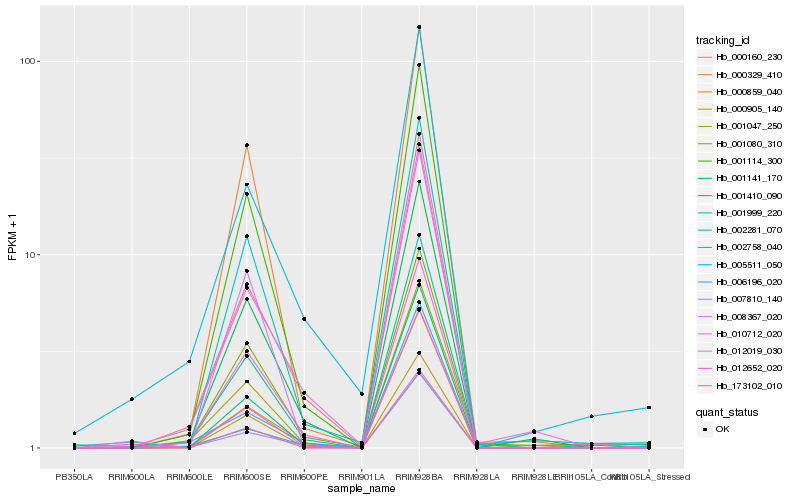
Hb_001999_220 |
0.0 |
- |
- |
PREDICTED: cytochrome P450 86A1 [Jatropha curcas] |
| 2 |
Hb_001410_090 |
0.0831350644 |
- |
- |
transferase, putative [Ricinus communis] |
| 3 |
Hb_001080_310 |
0.0839130785 |
- |
- |
PREDICTED: ent-copalyl diphosphate synthase, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000859_040 |
0.0855860542 |
- |
- |
Early nodulin 16 precursor, putative [Ricinus communis] |
| 5 |
Hb_001047_250 |
0.0874845425 |
- |
- |
PREDICTED: ent-copalyl diphosphate synthase, chloroplastic [Jatropha curcas] |
| 6 |
Hb_002281_070 |
0.0898008817 |
- |
- |
hypothetical protein JCGZ_04352 [Jatropha curcas] |
| 7 |
Hb_001114_300 |
0.0898851585 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 8 |
Hb_000160_230 |
0.0914677873 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 9 |
Hb_007810_140 |
0.091615734 |
- |
- |
Ankyrin repeat-containing protein [Medicago truncatula] |
| 10 |
Hb_002758_040 |
0.0951795152 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 11 |
Hb_005511_050 |
0.1006172833 |
- |
- |
hypothetical protein POPTR_0008s04130g [Populus trichocarpa] |
| 12 |
Hb_001141_170 |
0.1020482682 |
- |
- |
PREDICTED: non-specific lipid-transfer protein-like protein At2g13820 [Jatropha curcas] |
| 13 |
Hb_010712_020 |
0.103734636 |
- |
- |
PREDICTED: caffeic acid 3-O-methyltransferase-like [Jatropha curcas] |
| 14 |
Hb_006196_020 |
0.1054219144 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 100-like [Populus euphratica] |
| 15 |
Hb_012019_030 |
0.1061221678 |
- |
- |
hypothetical protein POPTR_0005s23360g [Populus trichocarpa] |
| 16 |
Hb_000329_410 |
0.1069314932 |
- |
- |
Beta-expansin 3 precursor, putative [Ricinus communis] |
| 17 |
Hb_000905_140 |
0.1075948015 |
- |
- |
PREDICTED: protein gamma response 1 isoform X2 [Jatropha curcas] |
| 18 |
Hb_008367_020 |
0.1125346703 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK1 [Gossypium raimondii] |
| 19 |
Hb_173102_010 |
0.1125955383 |
- |
- |
Ent-kaurene synthase B, chloroplast precursor, putative [Ricinus communis] |
| 20 |
Hb_012652_020 |
0.1128776435 |
- |
- |
PREDICTED: probable carboxylesterase 18 [Jatropha curcas] |