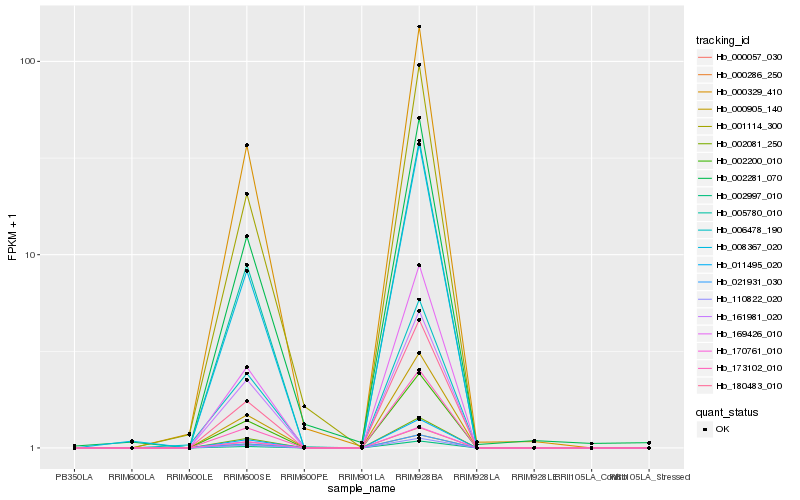
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000329_410 |
0.0 |
- |
- |
Beta-expansin 3 precursor, putative [Ricinus communis] |
| 2 |
Hb_000905_140 |
0.0353100044 |
- |
- |
PREDICTED: protein gamma response 1 isoform X2 [Jatropha curcas] |
| 3 |
Hb_011495_020 |
0.0415179589 |
- |
- |
Pectinesterase-2 precursor, putative [Ricinus communis] |
| 4 |
Hb_001114_300 |
0.044569389 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 5 |
Hb_000057_030 |
0.0457049937 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 6 |
Hb_002200_010 |
0.0459306851 |
- |
- |
PREDICTED: sperm acrosomal protein FSA-ACR.1-like [Jatropha curcas] |
| 7 |
Hb_180483_010 |
0.0462718849 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 8 |
Hb_005780_010 |
0.0465564736 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |
| 9 |
Hb_169426_010 |
0.0467009805 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 10 |
Hb_000286_250 |
0.0467584599 |
desease resistance |
Gene Name: NB-ARC |
- |
| 11 |
Hb_170761_010 |
0.0470537322 |
- |
- |
PREDICTED: uncharacterized protein LOC105787794 [Gossypium raimondii] |
| 12 |
Hb_002081_250 |
0.0471044749 |
- |
- |
hypothetical protein POPTR_0009s08940g [Populus trichocarpa] |
| 13 |
Hb_110822_020 |
0.0473906357 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 90-like [Jatropha curcas] |
| 14 |
Hb_002997_010 |
0.0502965033 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Malus domestica] |
| 15 |
Hb_021931_030 |
0.0549503482 |
- |
- |
hypothetical protein JCGZ_04974 [Jatropha curcas] |
| 16 |
Hb_006478_190 |
0.0560344362 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 17 |
Hb_161981_020 |
0.0585554339 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 18 |
Hb_008367_020 |
0.0587333506 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK1 [Gossypium raimondii] |
| 19 |
Hb_002281_070 |
0.0608171332 |
- |
- |
hypothetical protein JCGZ_04352 [Jatropha curcas] |
| 20 |
Hb_173102_010 |
0.0622656683 |
- |
- |
Ent-kaurene synthase B, chloroplast precursor, putative [Ricinus communis] |