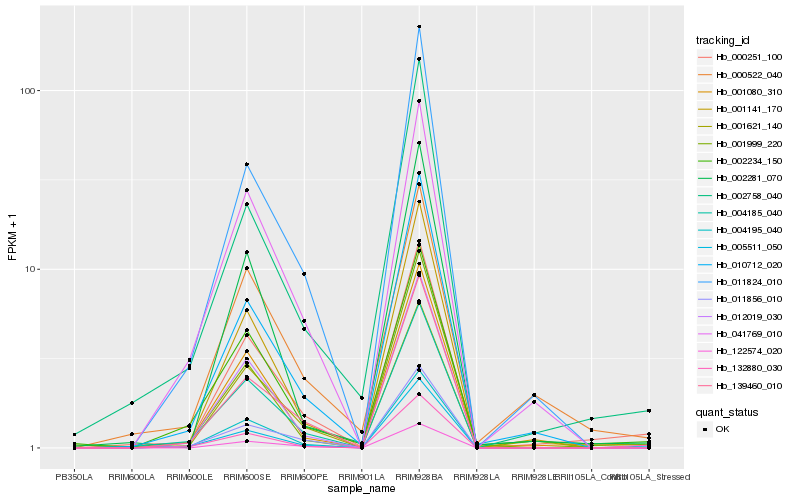
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000251_100 |
0.0 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 2 |
Hb_001080_310 |
0.0890102314 |
- |
- |
PREDICTED: ent-copalyl diphosphate synthase, chloroplastic [Jatropha curcas] |
| 3 |
Hb_005511_050 |
0.1054828611 |
- |
- |
hypothetical protein POPTR_0008s04130g [Populus trichocarpa] |
| 4 |
Hb_002758_040 |
0.1088386948 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 5 |
Hb_002234_150 |
0.1099097684 |
- |
- |
(S)-N-methylcoclaurine 3'-hydroxylase isozyme, putative [Ricinus communis] |
| 6 |
Hb_004185_040 |
0.1101533071 |
- |
- |
polygalacturonase, putative [Ricinus communis] |
| 7 |
Hb_004195_040 |
0.1130951021 |
- |
- |
salicylic acid methyl transferase [Capsicum annuum] |
| 8 |
Hb_001141_170 |
0.1160904585 |
- |
- |
PREDICTED: non-specific lipid-transfer protein-like protein At2g13820 [Jatropha curcas] |
| 9 |
Hb_132880_030 |
0.1165264369 |
- |
- |
hypothetical protein JCGZ_10325 [Jatropha curcas] |
| 10 |
Hb_012019_030 |
0.1168565481 |
- |
- |
hypothetical protein POPTR_0005s23360g [Populus trichocarpa] |
| 11 |
Hb_000522_040 |
0.1179288646 |
- |
- |
Desacetoxyvindoline 4-hydroxylase, putative [Ricinus communis] |
| 12 |
Hb_010712_020 |
0.1185242379 |
- |
- |
PREDICTED: caffeic acid 3-O-methyltransferase-like [Jatropha curcas] |
| 13 |
Hb_011824_010 |
0.1198653063 |
- |
- |
Metalloendoproteinase 1 precursor, putative [Ricinus communis] |
| 14 |
Hb_139460_010 |
0.1207506949 |
- |
- |
PREDICTED: beta-glucosidase 18-like [Jatropha curcas] |
| 15 |
Hb_122574_020 |
0.1209392633 |
- |
- |
hypothetical protein JCGZ_24076 [Jatropha curcas] |
| 16 |
Hb_001621_140 |
0.1215864781 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase 5 [Jatropha curcas] |
| 17 |
Hb_002281_070 |
0.1223110566 |
- |
- |
hypothetical protein JCGZ_04352 [Jatropha curcas] |
| 18 |
Hb_001999_220 |
0.1244796095 |
- |
- |
PREDICTED: cytochrome P450 86A1 [Jatropha curcas] |
| 19 |
Hb_011856_010 |
0.125936895 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 20 |
Hb_041769_010 |
0.1276571009 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |