| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
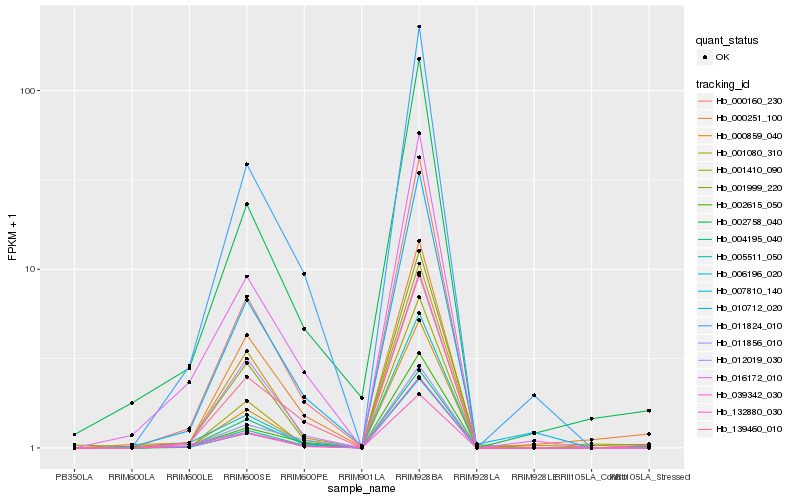
Hb_005511_050 |
0.0 |
- |
- |
hypothetical protein POPTR_0008s04130g [Populus trichocarpa] |
| 2 |
Hb_132880_030 |
0.0528381222 |
- |
- |
hypothetical protein JCGZ_10325 [Jatropha curcas] |
| 3 |
Hb_000160_230 |
0.0942670414 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 4 |
Hb_007810_140 |
0.0968960472 |
- |
- |
Ankyrin repeat-containing protein [Medicago truncatula] |
| 5 |
Hb_039342_030 |
0.0997056917 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001999_220 |
0.1006172833 |
- |
- |
PREDICTED: cytochrome P450 86A1 [Jatropha curcas] |
| 7 |
Hb_000859_040 |
0.1010796589 |
- |
- |
Early nodulin 16 precursor, putative [Ricinus communis] |
| 8 |
Hb_002758_040 |
0.1020961678 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 9 |
Hb_139460_010 |
0.1021396302 |
- |
- |
PREDICTED: beta-glucosidase 18-like [Jatropha curcas] |
| 10 |
Hb_002615_050 |
0.1026124599 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 11 |
Hb_011824_010 |
0.1026827292 |
- |
- |
Metalloendoproteinase 1 precursor, putative [Ricinus communis] |
| 12 |
Hb_016172_010 |
0.1051607592 |
- |
- |
hypothetical protein JCGZ_23602 [Jatropha curcas] |
| 13 |
Hb_012019_030 |
0.1053336771 |
- |
- |
hypothetical protein POPTR_0005s23360g [Populus trichocarpa] |
| 14 |
Hb_000251_100 |
0.1054828611 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 15 |
Hb_004195_040 |
0.1059263138 |
- |
- |
salicylic acid methyl transferase [Capsicum annuum] |
| 16 |
Hb_011856_010 |
0.1066343091 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 17 |
Hb_006196_020 |
0.1081773983 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 100-like [Populus euphratica] |
| 18 |
Hb_010712_020 |
0.1090789906 |
- |
- |
PREDICTED: caffeic acid 3-O-methyltransferase-like [Jatropha curcas] |
| 19 |
Hb_001410_090 |
0.1098855804 |
- |
- |
transferase, putative [Ricinus communis] |
| 20 |
Hb_001080_310 |
0.1106751378 |
- |
- |
PREDICTED: ent-copalyl diphosphate synthase, chloroplastic [Jatropha curcas] |