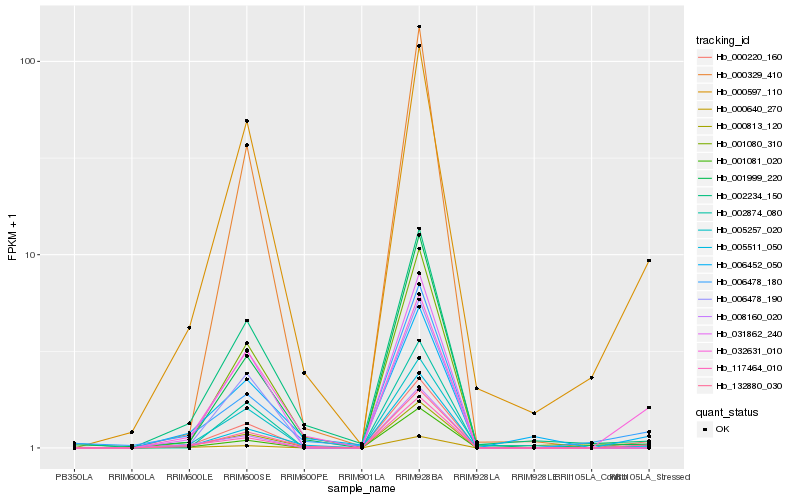
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000220_160 |
0.0 |
- |
- |
hypothetical protein B456_003G184800 [Gossypium raimondii] |
| 2 |
Hb_132880_030 |
0.0992306657 |
- |
- |
hypothetical protein JCGZ_10325 [Jatropha curcas] |
| 3 |
Hb_000597_110 |
0.1354023653 |
- |
- |
PREDICTED: VQ motif-containing protein 22-like [Jatropha curcas] |
| 4 |
Hb_032631_010 |
0.13716224 |
- |
- |
PREDICTED: uncharacterized protein LOC105638774 [Jatropha curcas] |
| 5 |
Hb_002874_080 |
0.1378312276 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 6 |
Hb_005511_050 |
0.1404842171 |
- |
- |
hypothetical protein POPTR_0008s04130g [Populus trichocarpa] |
| 7 |
Hb_031862_240 |
0.157827495 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 8 |
Hb_005257_020 |
0.1604007772 |
- |
- |
PREDICTED: ent-copalyl diphosphate synthase, chloroplastic [Jatropha curcas] |
| 9 |
Hb_006478_180 |
0.1612344172 |
- |
- |
Nonspecific lipid-transfer protein precursor, putative [Ricinus communis] |
| 10 |
Hb_006478_190 |
0.1651108054 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 11 |
Hb_006452_050 |
0.1678570667 |
- |
- |
PREDICTED: GABA transporter 1-like [Jatropha curcas] |
| 12 |
Hb_000640_270 |
0.1701960469 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: probable pectinesterase 30 [Jatropha curcas] |
| 13 |
Hb_001080_310 |
0.1701986544 |
- |
- |
PREDICTED: ent-copalyl diphosphate synthase, chloroplastic [Jatropha curcas] |
| 14 |
Hb_001081_020 |
0.1718441741 |
- |
- |
Cytochrome P450 76C2 [Triticum urartu] |
| 15 |
Hb_117464_010 |
0.1760255935 |
- |
- |
PREDICTED: flavone 3'-O-methyltransferase 1-like [Jatropha curcas] |
| 16 |
Hb_008160_020 |
0.1766710912 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 23-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_002234_150 |
0.1767757073 |
- |
- |
(S)-N-methylcoclaurine 3'-hydroxylase isozyme, putative [Ricinus communis] |
| 18 |
Hb_000813_120 |
0.1774313261 |
- |
- |
PREDICTED: serine carboxypeptidase-like 18 [Tarenaya hassleriana] |
| 19 |
Hb_001999_220 |
0.1779082727 |
- |
- |
PREDICTED: cytochrome P450 86A1 [Jatropha curcas] |
| 20 |
Hb_000329_410 |
0.1784294543 |
- |
- |
Beta-expansin 3 precursor, putative [Ricinus communis] |