| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
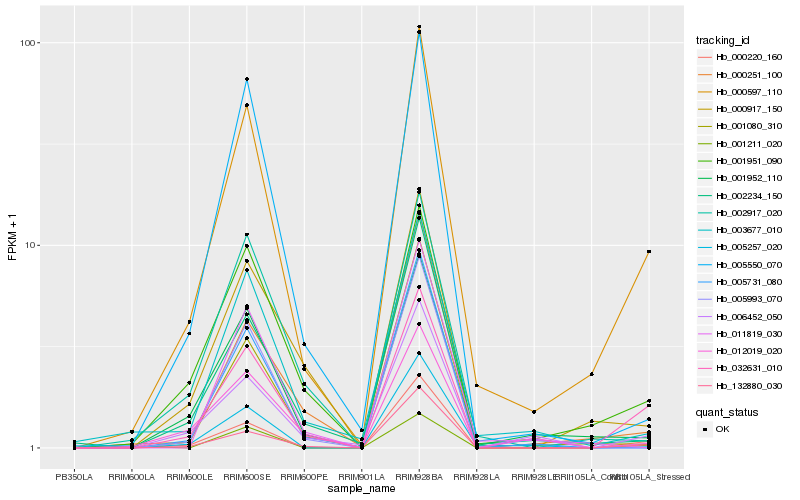
Hb_000597_110 |
0.0 |
- |
- |
PREDICTED: VQ motif-containing protein 22-like [Jatropha curcas] |
| 2 |
Hb_032631_010 |
0.1125516745 |
- |
- |
PREDICTED: uncharacterized protein LOC105638774 [Jatropha curcas] |
| 3 |
Hb_001952_110 |
0.1284147613 |
- |
- |
Ent-kaurene synthase B, chloroplast precursor, putative [Ricinus communis] |
| 4 |
Hb_000220_160 |
0.1354023653 |
- |
- |
hypothetical protein B456_003G184800 [Gossypium raimondii] |
| 5 |
Hb_001951_090 |
0.1382630922 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 6 |
Hb_003677_010 |
0.1505211004 |
transcription factor |
TF Family: SRS |
PREDICTED: protein SHI RELATED SEQUENCE 1 [Jatropha curcas] |
| 7 |
Hb_132880_030 |
0.1524478574 |
- |
- |
hypothetical protein JCGZ_10325 [Jatropha curcas] |
| 8 |
Hb_002234_150 |
0.155410784 |
- |
- |
(S)-N-methylcoclaurine 3'-hydroxylase isozyme, putative [Ricinus communis] |
| 9 |
Hb_006452_050 |
0.1587359172 |
- |
- |
PREDICTED: GABA transporter 1-like [Jatropha curcas] |
| 10 |
Hb_000251_100 |
0.1651220598 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 11 |
Hb_001080_310 |
0.1661117031 |
- |
- |
PREDICTED: ent-copalyl diphosphate synthase, chloroplastic [Jatropha curcas] |
| 12 |
Hb_005257_020 |
0.166344458 |
- |
- |
PREDICTED: ent-copalyl diphosphate synthase, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000917_150 |
0.1698065139 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_005550_070 |
0.1710180765 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 75 [Populus euphratica] |
| 15 |
Hb_002917_020 |
0.1782253638 |
- |
- |
predicted protein [Arabidopsis lyrata subsp. lyrata] |
| 16 |
Hb_012019_020 |
0.1808666236 |
- |
- |
Nonspecific lipid-transfer protein precursor, putative [Ricinus communis] |
| 17 |
Hb_001211_020 |
0.1821164508 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0002s11440g [Populus trichocarpa] |
| 18 |
Hb_005731_080 |
0.1824158045 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 19 |
Hb_005993_070 |
0.1843446238 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 20 |
Hb_011819_030 |
0.1849690541 |
- |
- |
cytochrome P450, putative [Ricinus communis] |