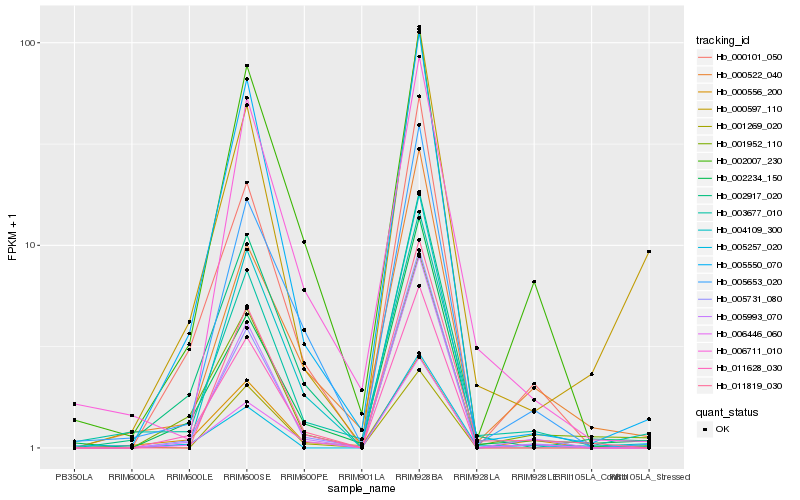
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003677_010 |
0.0 |
transcription factor |
TF Family: SRS |
PREDICTED: protein SHI RELATED SEQUENCE 1 [Jatropha curcas] |
| 2 |
Hb_006711_010 |
0.1185787534 |
- |
- |
cytoplasmic dynein light chain, putative [Ricinus communis] |
| 3 |
Hb_005550_070 |
0.1253475143 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 75 [Populus euphratica] |
| 4 |
Hb_005653_020 |
0.1271809589 |
transcription factor |
TF Family: C2H2 |
TRANSPARENT TESTA 1 protein, putative [Ricinus communis] |
| 5 |
Hb_000522_040 |
0.1284357035 |
- |
- |
Desacetoxyvindoline 4-hydroxylase, putative [Ricinus communis] |
| 6 |
Hb_002234_150 |
0.1324030409 |
- |
- |
(S)-N-methylcoclaurine 3'-hydroxylase isozyme, putative [Ricinus communis] |
| 7 |
Hb_001952_110 |
0.1350323159 |
- |
- |
Ent-kaurene synthase B, chloroplast precursor, putative [Ricinus communis] |
| 8 |
Hb_005993_070 |
0.1382206213 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 9 |
Hb_011819_030 |
0.1391146728 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 10 |
Hb_002917_020 |
0.1400690173 |
- |
- |
predicted protein [Arabidopsis lyrata subsp. lyrata] |
| 11 |
Hb_000556_200 |
0.1416511917 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 22 [Jatropha curcas] |
| 12 |
Hb_004109_300 |
0.1435539619 |
- |
- |
hypothetical protein POPTR_0003s21800g [Populus trichocarpa] |
| 13 |
Hb_006446_060 |
0.1435960203 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Populus euphratica] |
| 14 |
Hb_005731_080 |
0.1449937822 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 15 |
Hb_002007_230 |
0.1497278519 |
- |
- |
PREDICTED: probable linoleate 9S-lipoxygenase 5 [Jatropha curcas] |
| 16 |
Hb_001269_020 |
0.1497720529 |
- |
- |
PREDICTED: beta-Amyrin Synthase 2-like [Vitis vinifera] |
| 17 |
Hb_000597_110 |
0.1505211004 |
- |
- |
PREDICTED: VQ motif-containing protein 22-like [Jatropha curcas] |
| 18 |
Hb_005257_020 |
0.1518672269 |
- |
- |
PREDICTED: ent-copalyl diphosphate synthase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000101_050 |
0.1520455272 |
- |
- |
Metalloendoproteinase 1 precursor, putative [Ricinus communis] |
| 20 |
Hb_011628_030 |
0.1520887348 |
- |
- |
conserved hypothetical protein [Ricinus communis] |