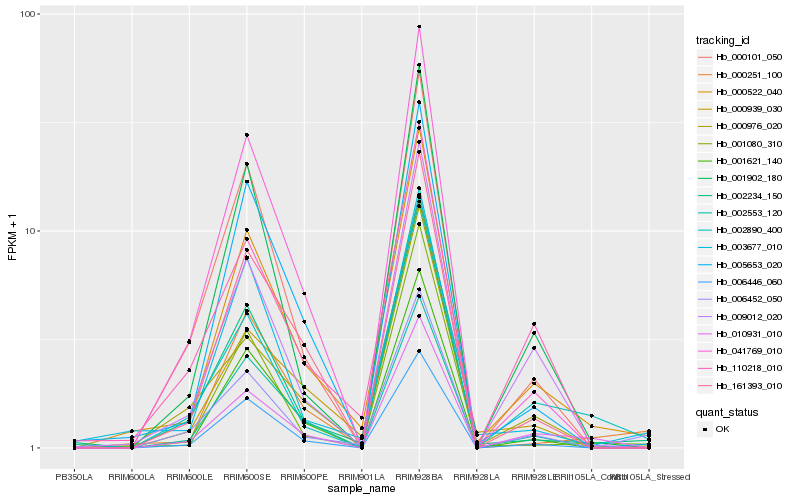
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000522_040 |
0.0 |
- |
- |
Desacetoxyvindoline 4-hydroxylase, putative [Ricinus communis] |
| 2 |
Hb_001621_140 |
0.1054986594 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase 5 [Jatropha curcas] |
| 3 |
Hb_005653_020 |
0.1114191787 |
transcription factor |
TF Family: C2H2 |
TRANSPARENT TESTA 1 protein, putative [Ricinus communis] |
| 4 |
Hb_006446_060 |
0.1154393889 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Populus euphratica] |
| 5 |
Hb_002234_150 |
0.1179198788 |
- |
- |
(S)-N-methylcoclaurine 3'-hydroxylase isozyme, putative [Ricinus communis] |
| 6 |
Hb_000251_100 |
0.1179288646 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 7 |
Hb_009012_020 |
0.1209752946 |
- |
- |
PREDICTED: 7-deoxyloganetic acid glucosyltransferase-like [Populus euphratica] |
| 8 |
Hb_010931_010 |
0.1211833777 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 9 |
Hb_161393_010 |
0.123140818 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 10 |
Hb_001902_180 |
0.1240171325 |
- |
- |
PREDICTED: uncharacterized protein LOC105129658 [Populus euphratica] |
| 11 |
Hb_041769_010 |
0.1257989688 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 12 |
Hb_003677_010 |
0.1284357035 |
transcription factor |
TF Family: SRS |
PREDICTED: protein SHI RELATED SEQUENCE 1 [Jatropha curcas] |
| 13 |
Hb_000976_020 |
0.1320348742 |
- |
- |
leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 14 |
Hb_000101_050 |
0.1327558144 |
- |
- |
Metalloendoproteinase 1 precursor, putative [Ricinus communis] |
| 15 |
Hb_002553_120 |
0.1345576752 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 1B [Jatropha curcas] |
| 16 |
Hb_000939_030 |
0.1356458839 |
- |
- |
PREDICTED: F-box protein At3g07870-like [Jatropha curcas] |
| 17 |
Hb_001080_310 |
0.1372128433 |
- |
- |
PREDICTED: ent-copalyl diphosphate synthase, chloroplastic [Jatropha curcas] |
| 18 |
Hb_110218_010 |
0.1378776031 |
- |
- |
PREDICTED: uncharacterized protein LOC104428128 [Eucalyptus grandis] |
| 19 |
Hb_002890_400 |
0.1383349274 |
- |
- |
2-nitropropane dioxygenase precursor, putative [Ricinus communis] |
| 20 |
Hb_006452_050 |
0.1408493956 |
- |
- |
PREDICTED: GABA transporter 1-like [Jatropha curcas] |