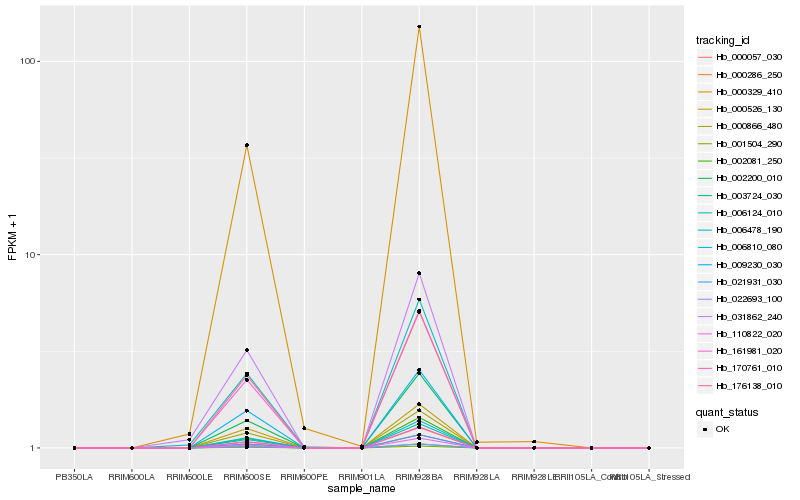
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_031862_240 |
0.0 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 2 |
Hb_006478_190 |
0.0422071597 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 3 |
Hb_006124_010 |
0.0745266081 |
- |
- |
- |
| 4 |
Hb_161981_020 |
0.0747952606 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 5 |
Hb_021931_030 |
0.075406946 |
- |
- |
hypothetical protein JCGZ_04974 [Jatropha curcas] |
| 6 |
Hb_176138_010 |
0.0755787591 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 22 [Jatropha curcas] |
| 7 |
Hb_006810_080 |
0.0756188143 |
- |
- |
hypothetical protein EUGRSUZ_C015472, partial [Eucalyptus grandis] |
| 8 |
Hb_022693_100 |
0.076610647 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 9 |
Hb_000866_480 |
0.0768177484 |
- |
- |
- |
| 10 |
Hb_110822_020 |
0.0783012241 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 90-like [Jatropha curcas] |
| 11 |
Hb_002081_250 |
0.0784734724 |
- |
- |
hypothetical protein POPTR_0009s08940g [Populus trichocarpa] |
| 12 |
Hb_170761_010 |
0.0785046987 |
- |
- |
PREDICTED: uncharacterized protein LOC105787794 [Gossypium raimondii] |
| 13 |
Hb_000286_250 |
0.0786906599 |
desease resistance |
Gene Name: NB-ARC |
- |
| 14 |
Hb_002200_010 |
0.0792541346 |
- |
- |
PREDICTED: sperm acrosomal protein FSA-ACR.1-like [Jatropha curcas] |
| 15 |
Hb_009230_030 |
0.0793573566 |
- |
- |
- |
| 16 |
Hb_000057_030 |
0.0794197149 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 17 |
Hb_000526_130 |
0.0821482424 |
- |
- |
PREDICTED: cytochrome P450 705A5-like [Jatropha curcas] |
| 18 |
Hb_003724_030 |
0.0829780374 |
- |
- |
- |
| 19 |
Hb_001504_290 |
0.0836318848 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 20 |
Hb_000329_410 |
0.084289804 |
- |
- |
Beta-expansin 3 precursor, putative [Ricinus communis] |