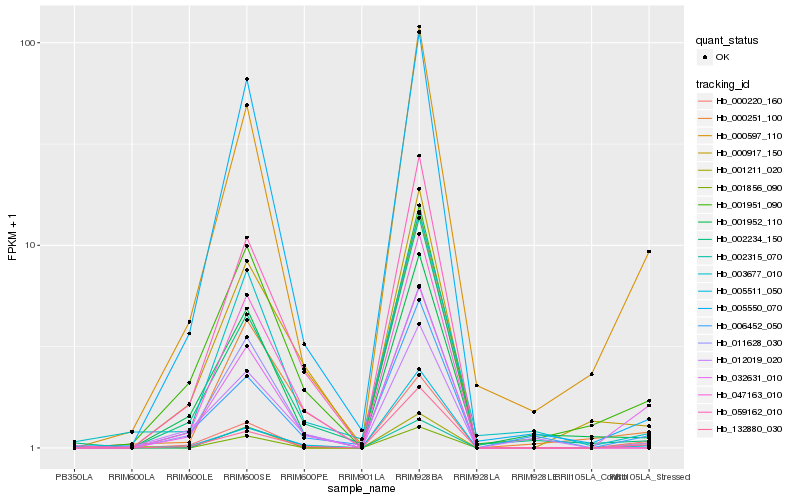
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_032631_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638774 [Jatropha curcas] |
| 2 |
Hb_000597_110 |
0.1125516745 |
- |
- |
PREDICTED: VQ motif-containing protein 22-like [Jatropha curcas] |
| 3 |
Hb_000220_160 |
0.13716224 |
- |
- |
hypothetical protein B456_003G184800 [Gossypium raimondii] |
| 4 |
Hb_132880_030 |
0.1481451537 |
- |
- |
hypothetical protein JCGZ_10325 [Jatropha curcas] |
| 5 |
Hb_001856_090 |
0.1534373169 |
- |
- |
Cellulose synthase-like B3, putative isoform 2 [Theobroma cacao] |
| 6 |
Hb_001951_090 |
0.1686393696 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 7 |
Hb_006452_050 |
0.1737102976 |
- |
- |
PREDICTED: GABA transporter 1-like [Jatropha curcas] |
| 8 |
Hb_000917_150 |
0.1836920457 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002315_070 |
0.1875187522 |
- |
- |
PREDICTED: uncharacterized protein LOC105630187 [Jatropha curcas] |
| 10 |
Hb_001211_020 |
0.1898383409 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0002s11440g [Populus trichocarpa] |
| 11 |
Hb_001952_110 |
0.1916266608 |
- |
- |
Ent-kaurene synthase B, chloroplast precursor, putative [Ricinus communis] |
| 12 |
Hb_005511_050 |
0.1928407305 |
- |
- |
hypothetical protein POPTR_0008s04130g [Populus trichocarpa] |
| 13 |
Hb_000251_100 |
0.1952456949 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 14 |
Hb_005550_070 |
0.1965372496 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 75 [Populus euphratica] |
| 15 |
Hb_011628_030 |
0.1971002221 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_012019_020 |
0.1971118297 |
- |
- |
Nonspecific lipid-transfer protein precursor, putative [Ricinus communis] |
| 17 |
Hb_047163_010 |
0.2002409016 |
- |
- |
PREDICTED: periaxin-like [Populus euphratica] |
| 18 |
Hb_002234_150 |
0.2006988232 |
- |
- |
(S)-N-methylcoclaurine 3'-hydroxylase isozyme, putative [Ricinus communis] |
| 19 |
Hb_059162_010 |
0.2014766137 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 20 |
Hb_003677_010 |
0.203236834 |
transcription factor |
TF Family: SRS |
PREDICTED: protein SHI RELATED SEQUENCE 1 [Jatropha curcas] |