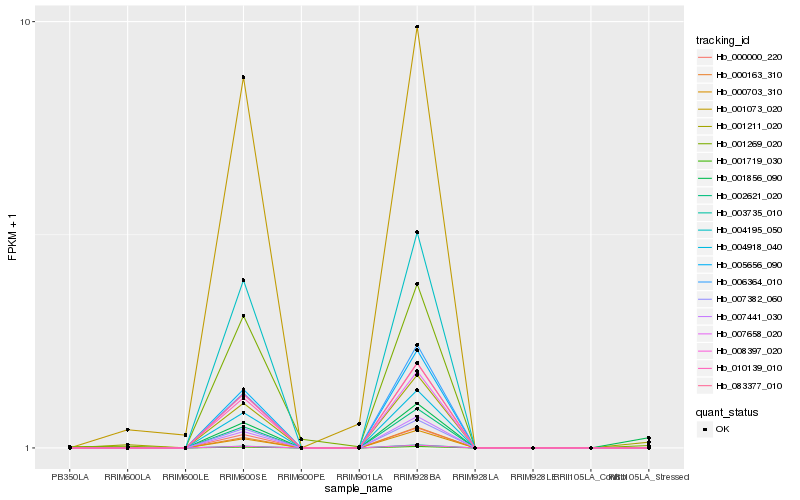
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001211_020 |
0.0 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0002s11440g [Populus trichocarpa] |
| 2 |
Hb_001856_090 |
0.1642750207 |
- |
- |
Cellulose synthase-like B3, putative isoform 2 [Theobroma cacao] |
| 3 |
Hb_001269_020 |
0.1760213654 |
- |
- |
PREDICTED: beta-Amyrin Synthase 2-like [Vitis vinifera] |
| 4 |
Hb_004918_040 |
0.1771017172 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_003735_010 |
0.1771465168 |
- |
- |
- |
| 6 |
Hb_008397_020 |
0.1771502843 |
- |
- |
polyprotein [Oryza australiensis] |
| 7 |
Hb_001719_030 |
0.1771713045 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 8 |
Hb_083377_010 |
0.1771930952 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 9 |
Hb_005656_090 |
0.1774370916 |
transcription factor |
TF Family: NAC |
hypothetical protein CICLE_v10008885mg [Citrus clementina] |
| 10 |
Hb_010139_010 |
0.1775010576 |
- |
- |
PREDICTED: uncharacterized protein LOC105645488 [Jatropha curcas] |
| 11 |
Hb_002621_020 |
0.1781066689 |
- |
- |
- |
| 12 |
Hb_000703_310 |
0.1784953914 |
- |
- |
- |
| 13 |
Hb_007441_030 |
0.1788648568 |
- |
- |
PREDICTED: uclacyanin-2-like [Jatropha curcas] |
| 14 |
Hb_007658_020 |
0.1796230203 |
- |
- |
Beta-amyrin synthase [Glycine soja] |
| 15 |
Hb_000163_310 |
0.179661417 |
- |
- |
PREDICTED: ABC transporter C family member 4-like [Malus domestica] |
| 16 |
Hb_000000_220 |
0.1796774312 |
- |
- |
protein with unknown function [Ricinus communis] |
| 17 |
Hb_007382_060 |
0.1803079399 |
- |
- |
- |
| 18 |
Hb_004195_050 |
0.1803153483 |
- |
- |
- |
| 19 |
Hb_001073_020 |
0.1808428585 |
- |
- |
hypothetical protein CISIN_1g031854mg [Citrus sinensis] |
| 20 |
Hb_006364_010 |
0.1814232129 |
- |
- |
- |