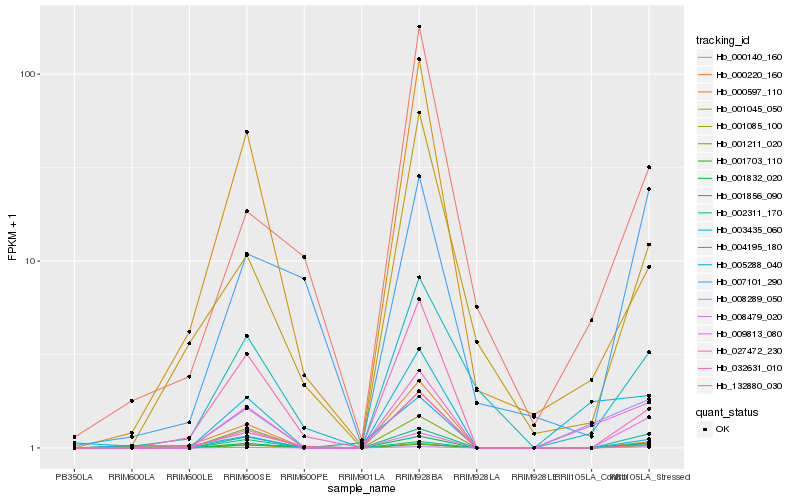
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001832_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104584029 [Brachypodium distachyon] |
| 2 |
Hb_001085_100 |
0.1113440479 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 3 |
Hb_008479_020 |
0.137415687 |
- |
- |
PREDICTED: uncharacterized protein LOC105436302 [Cucumis sativus] |
| 4 |
Hb_001856_090 |
0.1383975335 |
- |
- |
Cellulose synthase-like B3, putative isoform 2 [Theobroma cacao] |
| 5 |
Hb_027472_230 |
0.223607335 |
- |
- |
PREDICTED: auxin-induced protein 15A-like [Jatropha curcas] |
| 6 |
Hb_032631_010 |
0.2245787388 |
- |
- |
PREDICTED: uncharacterized protein LOC105638774 [Jatropha curcas] |
| 7 |
Hb_001045_050 |
0.2595021291 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0001s33560g [Populus trichocarpa] |
| 8 |
Hb_003435_060 |
0.2596496248 |
- |
- |
carbonic anhydrase, putative [Ricinus communis] |
| 9 |
Hb_000220_160 |
0.2598717531 |
- |
- |
hypothetical protein B456_003G184800 [Gossypium raimondii] |
| 10 |
Hb_000597_110 |
0.2718010651 |
- |
- |
PREDICTED: VQ motif-containing protein 22-like [Jatropha curcas] |
| 11 |
Hb_001211_020 |
0.2747459436 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0002s11440g [Populus trichocarpa] |
| 12 |
Hb_008289_050 |
0.2835948881 |
- |
- |
hypothetical protein B456_001G045900 [Gossypium raimondii] |
| 13 |
Hb_004195_180 |
0.2878289951 |
- |
- |
unnamed protein product [Coffea canephora] |
| 14 |
Hb_005288_040 |
0.2908478534 |
- |
- |
DUF26 domain-containing protein 2 precursor, putative [Ricinus communis] |
| 15 |
Hb_007101_290 |
0.2949499553 |
transcription factor |
TF Family: C2H2 |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000140_160 |
0.2961642344 |
- |
- |
PREDICTED: neurofilament light polypeptide-like [Gossypium raimondii] |
| 17 |
Hb_001703_110 |
0.2994033191 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Malus domestica] |
| 18 |
Hb_132880_030 |
0.3038129911 |
- |
- |
hypothetical protein JCGZ_10325 [Jatropha curcas] |
| 19 |
Hb_009813_080 |
0.304192637 |
- |
- |
hypothetical protein CISIN_1g045991mg [Citrus sinensis] |
| 20 |
Hb_002311_170 |
0.3104584543 |
- |
- |
PREDICTED: BTB/POZ and MATH domain-containing protein 2-like [Eucalyptus grandis] |