| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
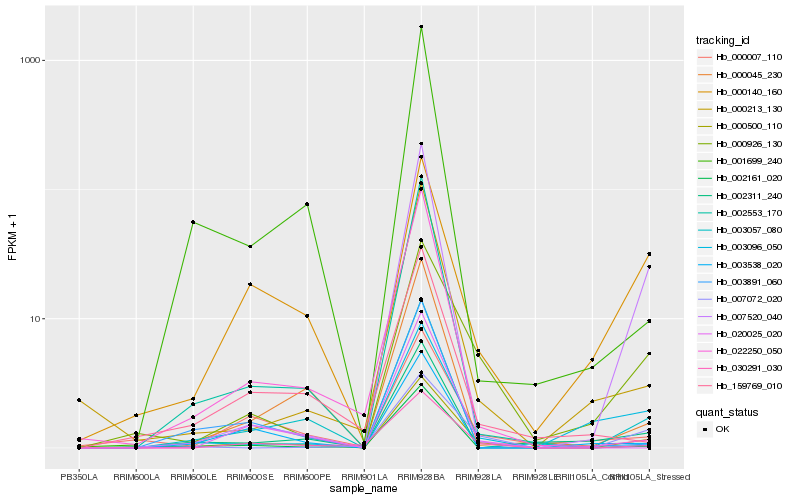
Hb_000500_110 |
0.0 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 2 |
Hb_003057_080 |
0.1550616827 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 1-like [Jatropha curcas] |
| 3 |
Hb_030291_030 |
0.1554168543 |
- |
- |
PREDICTED: agmatine coumaroyltransferase-2-like [Jatropha curcas] |
| 4 |
Hb_000926_130 |
0.1668805769 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 5 |
Hb_000045_230 |
0.1812369416 |
- |
- |
PREDICTED: uncharacterized acetyltransferase At3g50280 [Jatropha curcas] |
| 6 |
Hb_000140_160 |
0.1864083577 |
- |
- |
PREDICTED: neurofilament light polypeptide-like [Gossypium raimondii] |
| 7 |
Hb_007072_020 |
0.1870708047 |
transcription factor |
TF Family: bHLH |
lMYC5 [Hevea brasiliensis] |
| 8 |
Hb_000213_130 |
0.1933318401 |
transcription factor |
TF Family: HSF |
Heat Stress Transcription Factor family protein [Populus trichocarpa] |
| 9 |
Hb_020025_020 |
0.1935254919 |
- |
- |
SENESCENCE-RELATED GENE 1 family protein [Populus trichocarpa] |
| 10 |
Hb_002311_240 |
0.1968364937 |
- |
- |
hypothetical protein JCGZ_18937 [Jatropha curcas] |
| 11 |
Hb_002161_020 |
0.1974957304 |
transcription factor |
TF Family: bZIP |
hypothetical protein POPTR_0006s05770g, partial [Populus trichocarpa] |
| 12 |
Hb_022250_050 |
0.1995561829 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000007_110 |
0.1997881586 |
- |
- |
PREDICTED: peroxidase 11 [Jatropha curcas] |
| 14 |
Hb_007520_040 |
0.1998493764 |
- |
- |
PREDICTED: carbonic anhydrase 2-like [Jatropha curcas] |
| 15 |
Hb_003891_060 |
0.2027907703 |
- |
- |
PREDICTED: linoleate 13S-lipoxygenase 2-1, chloroplastic-like [Glycine max] |
| 16 |
Hb_003096_050 |
0.2035643743 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_159769_010 |
0.2042993571 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 18 |
Hb_003538_020 |
0.2046802136 |
- |
- |
(S)-N-methylcoclaurine 3'-hydroxylase isozyme, putative [Ricinus communis] |
| 19 |
Hb_001699_240 |
0.2051745733 |
- |
- |
PREDICTED: naringenin,2-oxoglutarate 3-dioxygenase [Jatropha curcas] |
| 20 |
Hb_002553_170 |
0.206223589 |
- |
- |
PREDICTED: early nodulin-75-like [Jatropha curcas] |