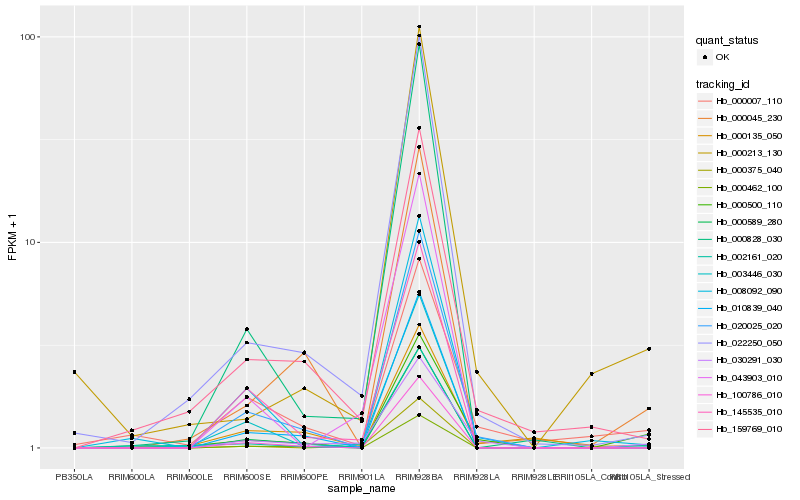
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_030291_030 |
0.0 |
- |
- |
PREDICTED: agmatine coumaroyltransferase-2-like [Jatropha curcas] |
| 2 |
Hb_000500_110 |
0.1554168543 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 3 |
Hb_020025_020 |
0.1760246937 |
- |
- |
SENESCENCE-RELATED GENE 1 family protein [Populus trichocarpa] |
| 4 |
Hb_159769_010 |
0.1772708859 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 5 |
Hb_022250_050 |
0.1807623445 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000007_110 |
0.1977378026 |
- |
- |
PREDICTED: peroxidase 11 [Jatropha curcas] |
| 7 |
Hb_003446_030 |
0.202500038 |
- |
- |
hypothetical protein JCGZ_20198 [Jatropha curcas] |
| 8 |
Hb_145535_010 |
0.2050208607 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |
| 9 |
Hb_002161_020 |
0.2057920561 |
transcription factor |
TF Family: bZIP |
hypothetical protein POPTR_0006s05770g, partial [Populus trichocarpa] |
| 10 |
Hb_043903_010 |
0.2077067046 |
- |
- |
PREDICTED: protein kinase 2B, chloroplastic-like [Tarenaya hassleriana] |
| 11 |
Hb_000045_230 |
0.2099224795 |
- |
- |
PREDICTED: uncharacterized acetyltransferase At3g50280 [Jatropha curcas] |
| 12 |
Hb_000375_040 |
0.2113201802 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-related protein 308-like [Populus euphratica] |
| 13 |
Hb_000213_130 |
0.211958247 |
transcription factor |
TF Family: HSF |
Heat Stress Transcription Factor family protein [Populus trichocarpa] |
| 14 |
Hb_000135_050 |
0.2128458159 |
- |
- |
PREDICTED: putative leucine-rich repeat receptor-like protein kinase At2g19210-like isoform X2 [Citrus sinensis] |
| 15 |
Hb_008092_090 |
0.2128773799 |
- |
- |
PREDICTED: uncharacterized protein LOC105637756 [Jatropha curcas] |
| 16 |
Hb_000589_280 |
0.2132707515 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase 5 [Jatropha curcas] |
| 17 |
Hb_000828_030 |
0.2141287472 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 18 |
Hb_000462_100 |
0.2143879524 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 19 |
Hb_010839_040 |
0.2144267846 |
- |
- |
hypothetical protein JCGZ_20868 [Jatropha curcas] |
| 20 |
Hb_100786_010 |
0.2151554434 |
- |
- |
hypothetical protein B456_008G202600 [Gossypium raimondii] |