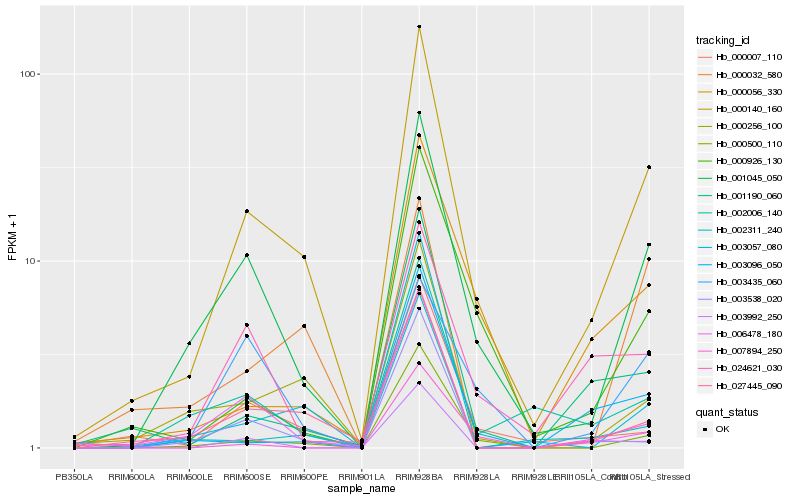
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000140_160 |
0.0 |
- |
- |
PREDICTED: neurofilament light polypeptide-like [Gossypium raimondii] |
| 2 |
Hb_001045_050 |
0.1394029077 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0001s33560g [Populus trichocarpa] |
| 3 |
Hb_027445_090 |
0.1839074393 |
- |
- |
oleosin 18.2 kDa-like [Jatropha curcas] |
| 4 |
Hb_000256_100 |
0.1850342351 |
transcription factor |
TF Family: ERF |
PPLZ02 family protein [Populus trichocarpa] |
| 5 |
Hb_000500_110 |
0.1864083577 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 6 |
Hb_001190_060 |
0.1918086574 |
transcription factor |
TF Family: GRAS |
PREDICTED: nodulation-signaling pathway 1 protein [Jatropha curcas] |
| 7 |
Hb_000007_110 |
0.1933590668 |
- |
- |
PREDICTED: peroxidase 11 [Jatropha curcas] |
| 8 |
Hb_002006_140 |
0.212052762 |
transcription factor |
TF Family: ERF |
hypothetical protein POPTR_0012s11030g [Populus trichocarpa] |
| 9 |
Hb_000926_130 |
0.2144534106 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 10 |
Hb_000056_330 |
0.2158957948 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 11 |
Hb_003096_050 |
0.2163654725 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_003057_080 |
0.2179043528 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 1-like [Jatropha curcas] |
| 13 |
Hb_007894_250 |
0.2229504248 |
- |
- |
hypothetical protein JCGZ_06575 [Jatropha curcas] |
| 14 |
Hb_024621_030 |
0.2269270778 |
- |
- |
tetraketide alpha-pyrone reductase 2 [Jatropha curcas] |
| 15 |
Hb_002311_240 |
0.2295449735 |
- |
- |
hypothetical protein JCGZ_18937 [Jatropha curcas] |
| 16 |
Hb_006478_180 |
0.2299961885 |
- |
- |
Nonspecific lipid-transfer protein precursor, putative [Ricinus communis] |
| 17 |
Hb_003538_020 |
0.2339347687 |
- |
- |
(S)-N-methylcoclaurine 3'-hydroxylase isozyme, putative [Ricinus communis] |
| 18 |
Hb_000032_580 |
0.2430452015 |
- |
- |
hypothetical protein RCOM_1311780 [Ricinus communis] |
| 19 |
Hb_003992_250 |
0.2435825782 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 2-like [Glycine max] |
| 20 |
Hb_003435_060 |
0.2437349486 |
- |
- |
carbonic anhydrase, putative [Ricinus communis] |