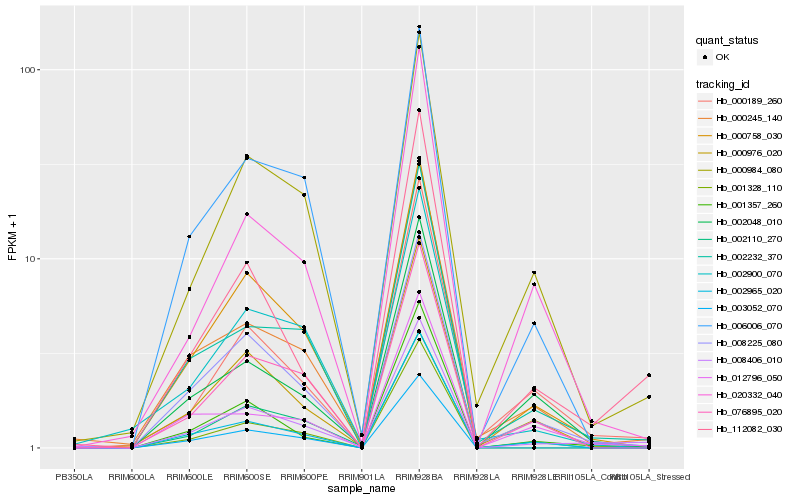
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008406_010 |
0.0 |
- |
- |
PREDICTED: tyrosine aminotransferase-like [Populus euphratica] |
| 2 |
Hb_008225_080 |
0.1120786651 |
- |
- |
PREDICTED: cytochrome P450 71A26-like [Jatropha curcas] |
| 3 |
Hb_002232_370 |
0.1126094174 |
- |
- |
PREDICTED: receptor-like protein kinase HAIKU2 [Jatropha curcas] |
| 4 |
Hb_000245_140 |
0.1139322259 |
- |
- |
PREDICTED: CASP-like protein 3A1 [Jatropha curcas] |
| 5 |
Hb_000758_030 |
0.1143564347 |
transcription factor |
TF Family: ERF |
Ethylene-responsive transcription factor, putative [Ricinus communis] |
| 6 |
Hb_112082_030 |
0.1151324354 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 7 |
Hb_001357_260 |
0.116524145 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 8 |
Hb_000976_020 |
0.1256436281 |
- |
- |
leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 9 |
Hb_000984_080 |
0.1354454917 |
- |
- |
PREDICTED: receptor-like protein kinase HAIKU2 [Jatropha curcas] |
| 10 |
Hb_002110_270 |
0.1364104331 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH96 [Jatropha curcas] |
| 11 |
Hb_000189_260 |
0.1374168304 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002965_020 |
0.1374358097 |
- |
- |
hypothetical protein JCGZ_18947 [Jatropha curcas] |
| 13 |
Hb_006006_070 |
0.1409914762 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 14 |
Hb_003052_070 |
0.1434295815 |
- |
- |
PREDICTED: peroxidase 11 [Jatropha curcas] |
| 15 |
Hb_020332_040 |
0.1462033029 |
- |
- |
PREDICTED: alcohol dehydrogenase 1-like [Jatropha curcas] |
| 16 |
Hb_076895_020 |
0.1467647566 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 isoform X2 [Jatropha curcas] |
| 17 |
Hb_002900_070 |
0.14683223 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002048_010 |
0.1468641833 |
- |
- |
unknown [Lotus japonicus] |
| 19 |
Hb_001328_110 |
0.1469280665 |
- |
- |
PREDICTED: LON peptidase N-terminal domain and RING finger protein 1-like [Jatropha curcas] |
| 20 |
Hb_012796_050 |
0.1469912819 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxyxylulose-5-phosphate synthase, putative [Ricinus communis] |