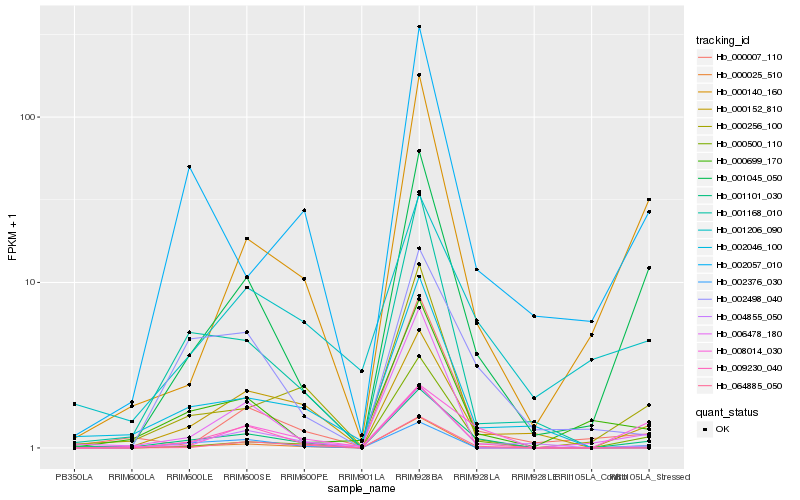
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001101_030 |
0.0 |
- |
- |
hypothetical protein JCGZ_00046 [Jatropha curcas] |
| 2 |
Hb_001045_050 |
0.1971551604 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0001s33560g [Populus trichocarpa] |
| 3 |
Hb_004855_050 |
0.2078963586 |
- |
- |
hypothetical protein POPTR_0016s12140g [Populus trichocarpa] |
| 4 |
Hb_000256_100 |
0.2178491257 |
transcription factor |
TF Family: ERF |
PPLZ02 family protein [Populus trichocarpa] |
| 5 |
Hb_009230_040 |
0.2212820862 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000152_810 |
0.2238372226 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 7 |
Hb_002498_040 |
0.2249454592 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 31 [Jatropha curcas] |
| 8 |
Hb_002046_100 |
0.2254297524 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB82 [Jatropha curcas] |
| 9 |
Hb_001206_090 |
0.2316713572 |
- |
- |
pyruvate decarboxylase [Hevea brasiliensis] |
| 10 |
Hb_000007_110 |
0.2324204504 |
- |
- |
PREDICTED: peroxidase 11 [Jatropha curcas] |
| 11 |
Hb_000699_170 |
0.2334940805 |
- |
- |
myosin XI, putative [Ricinus communis] |
| 12 |
Hb_008014_030 |
0.2356750124 |
- |
- |
Aquaporin NIP1.1, putative [Ricinus communis] |
| 13 |
Hb_000025_510 |
0.2368640197 |
- |
- |
PREDICTED: receptor-like protein 12 [Vitis vinifera] |
| 14 |
Hb_002057_010 |
0.2411947094 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 15 |
Hb_000140_160 |
0.2451147537 |
- |
- |
PREDICTED: neurofilament light polypeptide-like [Gossypium raimondii] |
| 16 |
Hb_002376_030 |
0.2453158386 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 17 |
Hb_000500_110 |
0.2477961329 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 18 |
Hb_064885_050 |
0.2484315906 |
- |
- |
PREDICTED: BAHD acyltransferase At5g47980-like [Populus euphratica] |
| 19 |
Hb_006478_180 |
0.249298403 |
- |
- |
Nonspecific lipid-transfer protein precursor, putative [Ricinus communis] |
| 20 |
Hb_001168_010 |
0.2497682748 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 1 [Jatropha curcas] |