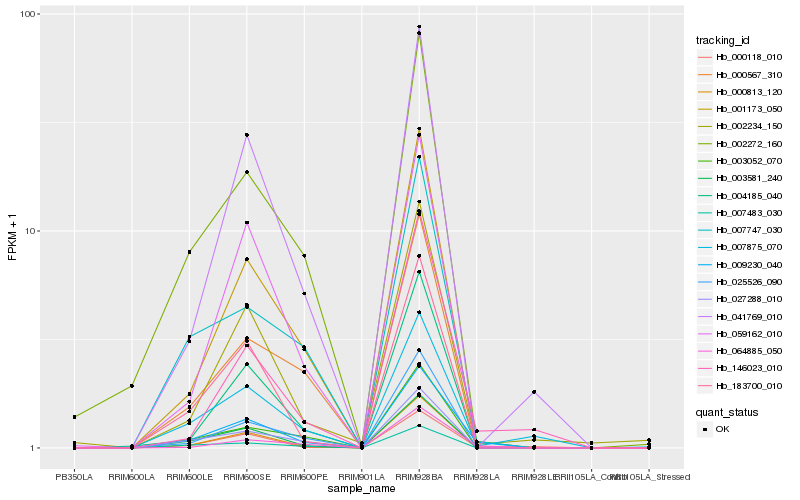
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009230_040 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_064885_050 |
0.1321984264 |
- |
- |
PREDICTED: BAHD acyltransferase At5g47980-like [Populus euphratica] |
| 3 |
Hb_003581_240 |
0.1384007343 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 4 |
Hb_000813_120 |
0.1393782624 |
- |
- |
PREDICTED: serine carboxypeptidase-like 18 [Tarenaya hassleriana] |
| 5 |
Hb_000118_010 |
0.1448548271 |
desease resistance |
Gene Name: LRR_4 |
hypothetical protein POPTR_0021s00470g [Populus trichocarpa] |
| 6 |
Hb_027288_010 |
0.1452096591 |
- |
- |
mutt domain protein, putative [Ricinus communis] |
| 7 |
Hb_001173_050 |
0.1456481671 |
- |
- |
DUF26 domain-containing protein 2 precursor, putative [Ricinus communis] |
| 8 |
Hb_007483_030 |
0.1516460566 |
- |
- |
- |
| 9 |
Hb_183700_010 |
0.151924148 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 10 |
Hb_007747_030 |
0.1549523015 |
- |
- |
PREDICTED: WD repeat-containing protein LWD2-like [Jatropha curcas] |
| 11 |
Hb_041769_010 |
0.1571997502 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 12 |
Hb_003052_070 |
0.1572259155 |
- |
- |
PREDICTED: peroxidase 11 [Jatropha curcas] |
| 13 |
Hb_004185_040 |
0.1577323508 |
- |
- |
polygalacturonase, putative [Ricinus communis] |
| 14 |
Hb_002272_160 |
0.1586518226 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 15 |
Hb_025526_090 |
0.1587989973 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 [Jatropha curcas] |
| 16 |
Hb_146023_010 |
0.1589347215 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 17 |
Hb_059162_010 |
0.158945718 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 18 |
Hb_002234_150 |
0.1605332567 |
- |
- |
(S)-N-methylcoclaurine 3'-hydroxylase isozyme, putative [Ricinus communis] |
| 19 |
Hb_007875_070 |
0.1609918118 |
- |
- |
hypothetical protein JCGZ_05017 [Jatropha curcas] |
| 20 |
Hb_000567_310 |
0.1628876712 |
- |
- |
Miraculin precursor, putative [Ricinus communis] |