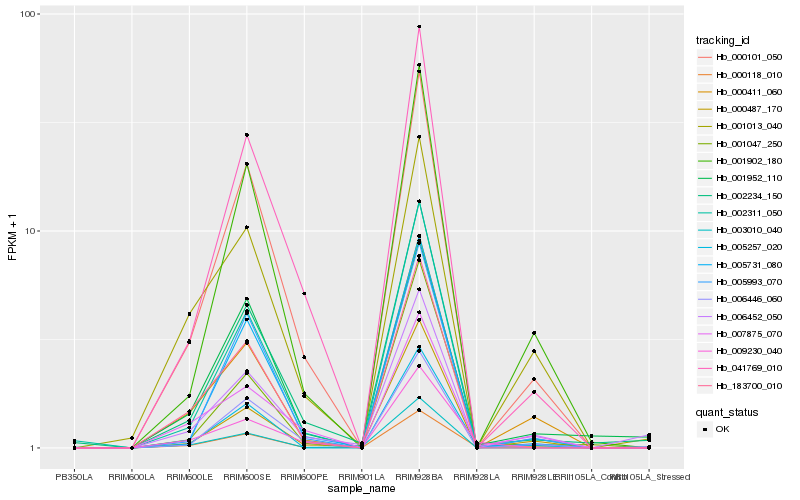
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000118_010 |
0.0 |
desease resistance |
Gene Name: LRR_4 |
hypothetical protein POPTR_0021s00470g [Populus trichocarpa] |
| 2 |
Hb_000101_050 |
0.1186886687 |
- |
- |
Metalloendoproteinase 1 precursor, putative [Ricinus communis] |
| 3 |
Hb_005257_020 |
0.1193955981 |
- |
- |
PREDICTED: ent-copalyl diphosphate synthase, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000411_060 |
0.1221635491 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 5 |
Hb_002234_150 |
0.1328342631 |
- |
- |
(S)-N-methylcoclaurine 3'-hydroxylase isozyme, putative [Ricinus communis] |
| 6 |
Hb_001902_180 |
0.1357577931 |
- |
- |
PREDICTED: uncharacterized protein LOC105129658 [Populus euphratica] |
| 7 |
Hb_183700_010 |
0.1432498094 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 8 |
Hb_007875_070 |
0.1439731068 |
- |
- |
hypothetical protein JCGZ_05017 [Jatropha curcas] |
| 9 |
Hb_009230_040 |
0.1448548271 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002311_050 |
0.1452377344 |
- |
- |
PREDICTED: uncharacterized protein LOC105122837 isoform X2 [Populus euphratica] |
| 11 |
Hb_005731_080 |
0.1457571824 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 12 |
Hb_006446_060 |
0.1458886944 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Populus euphratica] |
| 13 |
Hb_000487_170 |
0.1479055165 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 14 |
Hb_005993_070 |
0.1489008434 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 15 |
Hb_001952_110 |
0.1489272153 |
- |
- |
Ent-kaurene synthase B, chloroplast precursor, putative [Ricinus communis] |
| 16 |
Hb_041769_010 |
0.1492489908 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 17 |
Hb_003010_040 |
0.1500985493 |
- |
- |
hypothetical protein AALP_AA7G248200 [Arabis alpina] |
| 18 |
Hb_001047_250 |
0.1529914592 |
- |
- |
PREDICTED: ent-copalyl diphosphate synthase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_001013_040 |
0.153139701 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 51 [Jatropha curcas] |
| 20 |
Hb_006452_050 |
0.1532225784 |
- |
- |
PREDICTED: GABA transporter 1-like [Jatropha curcas] |