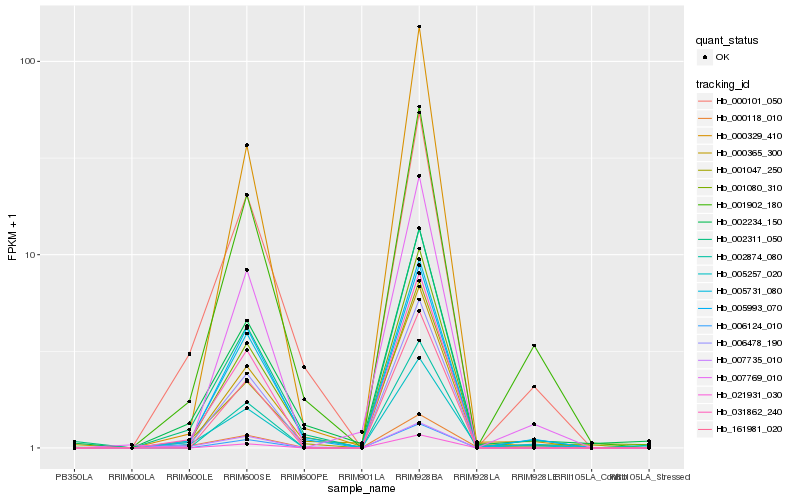
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005257_020 |
0.0 |
- |
- |
PREDICTED: ent-copalyl diphosphate synthase, chloroplastic [Jatropha curcas] |
| 2 |
Hb_002311_050 |
0.1181028455 |
- |
- |
PREDICTED: uncharacterized protein LOC105122837 isoform X2 [Populus euphratica] |
| 3 |
Hb_031862_240 |
0.1182097785 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 4 |
Hb_000118_010 |
0.1193955981 |
desease resistance |
Gene Name: LRR_4 |
hypothetical protein POPTR_0021s00470g [Populus trichocarpa] |
| 5 |
Hb_002234_150 |
0.1221258439 |
- |
- |
(S)-N-methylcoclaurine 3'-hydroxylase isozyme, putative [Ricinus communis] |
| 6 |
Hb_001080_310 |
0.1242895261 |
- |
- |
PREDICTED: ent-copalyl diphosphate synthase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_006478_190 |
0.1291067991 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 8 |
Hb_005731_080 |
0.129965767 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 9 |
Hb_005993_070 |
0.1306434777 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 10 |
Hb_001047_250 |
0.1323626596 |
- |
- |
PREDICTED: ent-copalyl diphosphate synthase, chloroplastic [Jatropha curcas] |
| 11 |
Hb_001902_180 |
0.1343070074 |
- |
- |
PREDICTED: uncharacterized protein LOC105129658 [Populus euphratica] |
| 12 |
Hb_007735_010 |
0.1384669787 |
- |
- |
Gibberellin 20 oxidase, putative [Ricinus communis] |
| 13 |
Hb_000365_300 |
0.1414426298 |
- |
- |
PREDICTED: cytochrome P450 714C2-like [Jatropha curcas] |
| 14 |
Hb_002874_080 |
0.1428750564 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 15 |
Hb_007769_010 |
0.1430427447 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 16 |
Hb_000329_410 |
0.14336068 |
- |
- |
Beta-expansin 3 precursor, putative [Ricinus communis] |
| 17 |
Hb_000101_050 |
0.1437938464 |
- |
- |
Metalloendoproteinase 1 precursor, putative [Ricinus communis] |
| 18 |
Hb_006124_010 |
0.1479429394 |
- |
- |
- |
| 19 |
Hb_161981_020 |
0.14801855 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 20 |
Hb_021931_030 |
0.1482792049 |
- |
- |
hypothetical protein JCGZ_04974 [Jatropha curcas] |