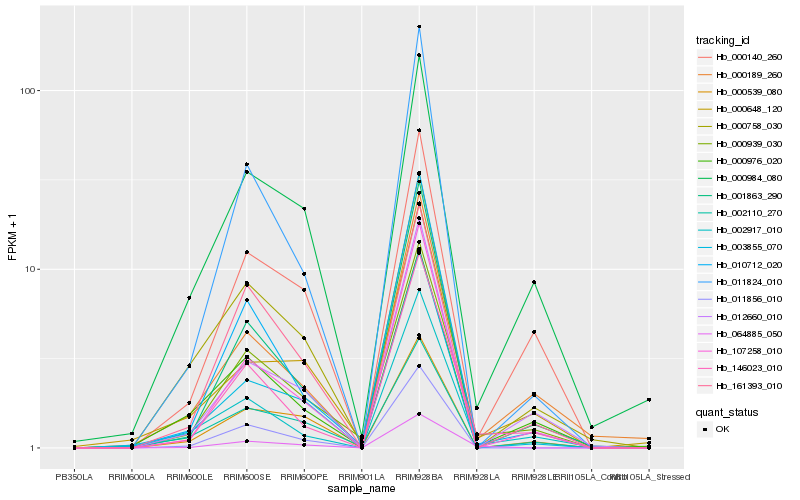
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000939_030 |
0.0 |
- |
- |
PREDICTED: F-box protein At3g07870-like [Jatropha curcas] |
| 2 |
Hb_146023_010 |
0.0945996027 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 3 |
Hb_000976_020 |
0.1131694445 |
- |
- |
leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 4 |
Hb_010712_020 |
0.1138450185 |
- |
- |
PREDICTED: caffeic acid 3-O-methyltransferase-like [Jatropha curcas] |
| 5 |
Hb_107258_010 |
0.1171694713 |
- |
- |
hypothetical protein JCGZ_01141 [Jatropha curcas] |
| 6 |
Hb_000140_260 |
0.1187390919 |
- |
- |
PREDICTED: uncharacterized protein LOC105642920 [Jatropha curcas] |
| 7 |
Hb_002110_270 |
0.1207351379 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH96 [Jatropha curcas] |
| 8 |
Hb_011824_010 |
0.1233932814 |
- |
- |
Metalloendoproteinase 1 precursor, putative [Ricinus communis] |
| 9 |
Hb_002917_010 |
0.1253291214 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1 [Jatropha curcas] |
| 10 |
Hb_012660_010 |
0.1257391713 |
- |
- |
hypothetical protein JCGZ_25971 [Jatropha curcas] |
| 11 |
Hb_000189_260 |
0.1262341699 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001863_290 |
0.1264012133 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Jatropha curcas] |
| 13 |
Hb_064885_050 |
0.1287815229 |
- |
- |
PREDICTED: BAHD acyltransferase At5g47980-like [Populus euphratica] |
| 14 |
Hb_000758_030 |
0.1288823391 |
transcription factor |
TF Family: ERF |
Ethylene-responsive transcription factor, putative [Ricinus communis] |
| 15 |
Hb_000648_120 |
0.1294856082 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase IV.1 [Vitis vinifera] |
| 16 |
Hb_161393_010 |
0.1296442081 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 17 |
Hb_000984_080 |
0.1307248447 |
- |
- |
PREDICTED: receptor-like protein kinase HAIKU2 [Jatropha curcas] |
| 18 |
Hb_011856_010 |
0.1307406555 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 19 |
Hb_003855_070 |
0.1319490344 |
- |
- |
hypothetical protein POPTR_0001s30980g [Populus trichocarpa] |
| 20 |
Hb_000539_080 |
0.1331601657 |
- |
- |
hypothetical protein POPTR_0039s00260g, partial [Populus trichocarpa] |