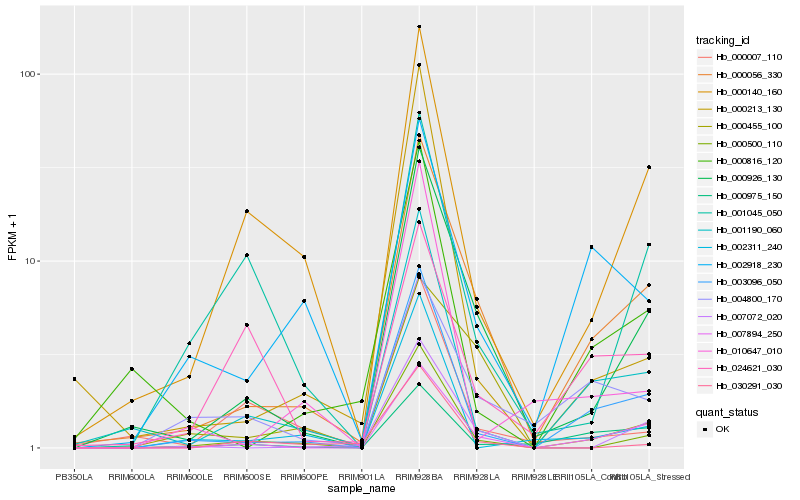
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000056_330 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_000926_130 |
0.1137719714 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 3 |
Hb_007894_250 |
0.1204305714 |
- |
- |
hypothetical protein JCGZ_06575 [Jatropha curcas] |
| 4 |
Hb_003096_050 |
0.1496095784 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001190_060 |
0.1804788733 |
transcription factor |
TF Family: GRAS |
PREDICTED: nodulation-signaling pathway 1 protein [Jatropha curcas] |
| 6 |
Hb_007072_020 |
0.1889829867 |
transcription factor |
TF Family: bHLH |
lMYC5 [Hevea brasiliensis] |
| 7 |
Hb_000975_150 |
0.2116098461 |
- |
- |
PREDICTED: phosphatidylinositol/phosphatidylcholine transfer protein SFH8 isoform X1 [Jatropha curcas] |
| 8 |
Hb_004800_170 |
0.2151056379 |
- |
- |
PREDICTED: cytochrome P450 90B1 [Jatropha curcas] |
| 9 |
Hb_000500_110 |
0.2153522463 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 10 |
Hb_000140_160 |
0.2158957948 |
- |
- |
PREDICTED: neurofilament light polypeptide-like [Gossypium raimondii] |
| 11 |
Hb_002918_230 |
0.2167891894 |
- |
- |
PREDICTED: probable glutathione S-transferase [Gossypium raimondii] |
| 12 |
Hb_000816_120 |
0.2171208659 |
- |
- |
PREDICTED: uncharacterized protein LOC105649214 [Jatropha curcas] |
| 13 |
Hb_000455_100 |
0.2369673535 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 14 |
Hb_000007_110 |
0.2627555801 |
- |
- |
PREDICTED: peroxidase 11 [Jatropha curcas] |
| 15 |
Hb_024621_030 |
0.2630476564 |
- |
- |
tetraketide alpha-pyrone reductase 2 [Jatropha curcas] |
| 16 |
Hb_001045_050 |
0.2632859404 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0001s33560g [Populus trichocarpa] |
| 17 |
Hb_000213_130 |
0.264576797 |
transcription factor |
TF Family: HSF |
Heat Stress Transcription Factor family protein [Populus trichocarpa] |
| 18 |
Hb_002311_240 |
0.2650549956 |
- |
- |
hypothetical protein JCGZ_18937 [Jatropha curcas] |
| 19 |
Hb_010647_010 |
0.2730186003 |
- |
- |
PREDICTED: protein NP24-like [Eucalyptus grandis] |
| 20 |
Hb_030291_030 |
0.2750886433 |
- |
- |
PREDICTED: agmatine coumaroyltransferase-2-like [Jatropha curcas] |