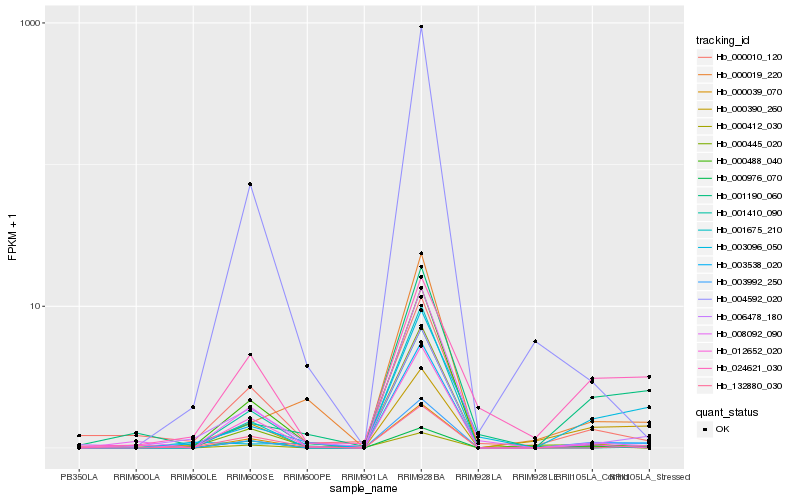
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003992_250 |
0.0 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 2-like [Glycine max] |
| 2 |
Hb_000039_070 |
0.168326643 |
transcription factor |
TF Family: MYB |
MYB21, putative [Theobroma cacao] |
| 3 |
Hb_003538_020 |
0.1849766693 |
- |
- |
(S)-N-methylcoclaurine 3'-hydroxylase isozyme, putative [Ricinus communis] |
| 4 |
Hb_001190_060 |
0.1859571857 |
transcription factor |
TF Family: GRAS |
PREDICTED: nodulation-signaling pathway 1 protein [Jatropha curcas] |
| 5 |
Hb_000976_070 |
0.1861524278 |
- |
- |
gulonolactone oxidase, putative [Ricinus communis] |
| 6 |
Hb_000412_030 |
0.2009516118 |
- |
- |
glycosyl hydrolase family 35 family protein [Populus trichocarpa] |
| 7 |
Hb_000390_260 |
0.2023286528 |
transcription factor |
TF Family: MYB |
Myb domain protein 107 [Theobroma cacao] |
| 8 |
Hb_006478_180 |
0.2096576247 |
- |
- |
Nonspecific lipid-transfer protein precursor, putative [Ricinus communis] |
| 9 |
Hb_000445_020 |
0.2194355937 |
- |
- |
GABA-specific permease, putative [Ricinus communis] |
| 10 |
Hb_012652_020 |
0.2226739248 |
- |
- |
PREDICTED: probable carboxylesterase 18 [Jatropha curcas] |
| 11 |
Hb_008092_090 |
0.2230055256 |
- |
- |
PREDICTED: uncharacterized protein LOC105637756 [Jatropha curcas] |
| 12 |
Hb_003096_050 |
0.2248689653 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000010_120 |
0.2273510614 |
- |
- |
PREDICTED: cytochrome P450 71A26-like [Jatropha curcas] |
| 14 |
Hb_024621_030 |
0.227966547 |
- |
- |
tetraketide alpha-pyrone reductase 2 [Jatropha curcas] |
| 15 |
Hb_001675_210 |
0.2290444453 |
- |
- |
PREDICTED: protein PLANT CADMIUM RESISTANCE 8-like [Jatropha curcas] |
| 16 |
Hb_132880_030 |
0.2374817287 |
- |
- |
hypothetical protein JCGZ_10325 [Jatropha curcas] |
| 17 |
Hb_000019_220 |
0.2375000029 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF113-like [Jatropha curcas] |
| 18 |
Hb_001410_090 |
0.2381996099 |
- |
- |
transferase, putative [Ricinus communis] |
| 19 |
Hb_000488_040 |
0.238307793 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 20 |
Hb_004592_020 |
0.2395993699 |
- |
- |
PREDICTED: non-specific lipid-transfer protein 1-like [Jatropha curcas] |