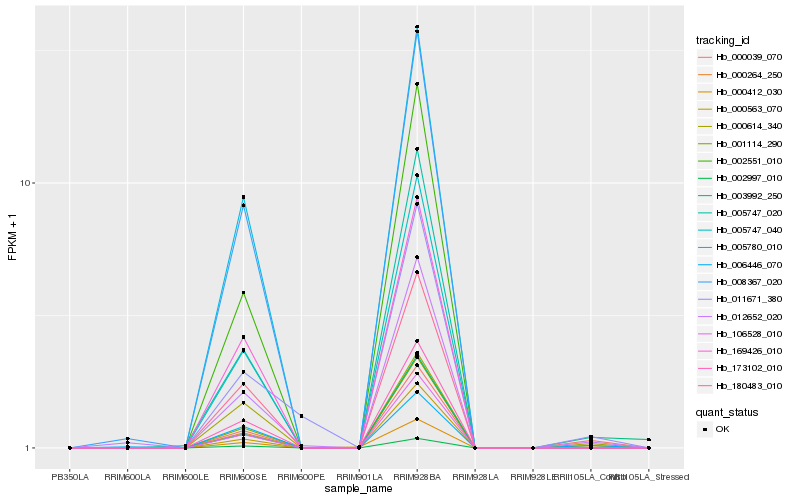
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000039_070 |
0.0 |
transcription factor |
TF Family: MYB |
MYB21, putative [Theobroma cacao] |
| 2 |
Hb_000412_030 |
0.1060943224 |
- |
- |
glycosyl hydrolase family 35 family protein [Populus trichocarpa] |
| 3 |
Hb_012652_020 |
0.1335493415 |
- |
- |
PREDICTED: probable carboxylesterase 18 [Jatropha curcas] |
| 4 |
Hb_006446_070 |
0.1530885658 |
- |
- |
Zinc-binding dehydrogenase family protein [Theobroma cacao] |
| 5 |
Hb_000614_340 |
0.1616146026 |
- |
- |
Salutaridinol 7-O-acetyltransferase, putative [Ricinus communis] |
| 6 |
Hb_001114_290 |
0.1637878706 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_005747_040 |
0.1646445404 |
- |
- |
PREDICTED: peroxidase N1-like, partial [Nicotiana sylvestris] |
| 8 |
Hb_106528_010 |
0.1648041345 |
- |
- |
Cationic peroxidase 2 precursor, putative [Ricinus communis] |
| 9 |
Hb_173102_010 |
0.1649902227 |
- |
- |
Ent-kaurene synthase B, chloroplast precursor, putative [Ricinus communis] |
| 10 |
Hb_002551_010 |
0.1661395922 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 11 |
Hb_002997_010 |
0.1679544174 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Malus domestica] |
| 12 |
Hb_000264_250 |
0.1682726946 |
- |
- |
thioredoxin family protein [Arabidopsis thaliana] |
| 13 |
Hb_003992_250 |
0.168326643 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 2-like [Glycine max] |
| 14 |
Hb_169426_010 |
0.1695095812 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 15 |
Hb_005780_010 |
0.1695840141 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |
| 16 |
Hb_180483_010 |
0.1697341068 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 17 |
Hb_011671_380 |
0.1704124608 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 18 |
Hb_008367_020 |
0.1708634224 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK1 [Gossypium raimondii] |
| 19 |
Hb_000563_070 |
0.1709612032 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase LOG2 [Sesamum indicum] |
| 20 |
Hb_005747_020 |
0.1724145765 |
- |
- |
bacterial-induced peroxidase [Gossypium schwendimanii] |