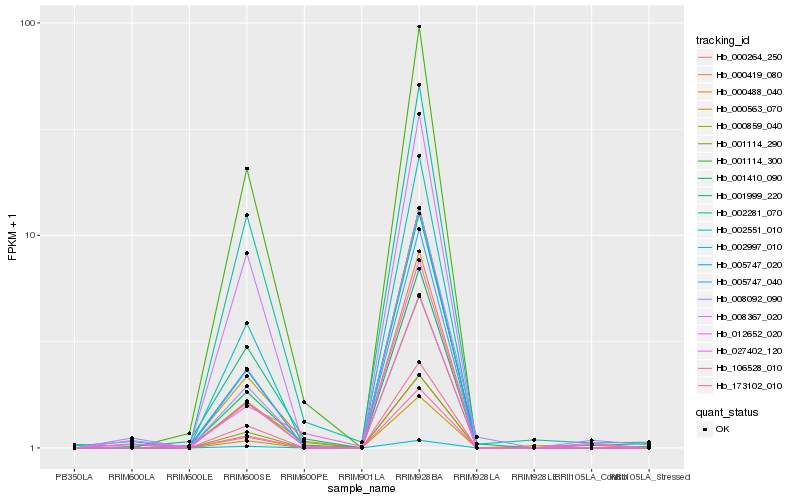
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012652_020 |
0.0 |
- |
- |
PREDICTED: probable carboxylesterase 18 [Jatropha curcas] |
| 2 |
Hb_008367_020 |
0.1064429907 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK1 [Gossypium raimondii] |
| 3 |
Hb_001410_090 |
0.1078058141 |
- |
- |
transferase, putative [Ricinus communis] |
| 4 |
Hb_000488_040 |
0.1099409039 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 5 |
Hb_001114_290 |
0.1100342869 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_005747_040 |
0.1100937908 |
- |
- |
PREDICTED: peroxidase N1-like, partial [Nicotiana sylvestris] |
| 7 |
Hb_106528_010 |
0.1102373167 |
- |
- |
Cationic peroxidase 2 precursor, putative [Ricinus communis] |
| 8 |
Hb_002551_010 |
0.1116321175 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 9 |
Hb_001999_220 |
0.1128776435 |
- |
- |
PREDICTED: cytochrome P450 86A1 [Jatropha curcas] |
| 10 |
Hb_173102_010 |
0.1130189768 |
- |
- |
Ent-kaurene synthase B, chloroplast precursor, putative [Ricinus communis] |
| 11 |
Hb_002281_070 |
0.1138591345 |
- |
- |
hypothetical protein JCGZ_04352 [Jatropha curcas] |
| 12 |
Hb_000264_250 |
0.1141477205 |
- |
- |
thioredoxin family protein [Arabidopsis thaliana] |
| 13 |
Hb_000859_040 |
0.1145101844 |
- |
- |
Early nodulin 16 precursor, putative [Ricinus communis] |
| 14 |
Hb_008092_090 |
0.115508897 |
- |
- |
PREDICTED: uncharacterized protein LOC105637756 [Jatropha curcas] |
| 15 |
Hb_001114_300 |
0.1157316873 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 16 |
Hb_027402_120 |
0.1167116601 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 17 |
Hb_000563_070 |
0.1174908011 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase LOG2 [Sesamum indicum] |
| 18 |
Hb_002997_010 |
0.1184334741 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Malus domestica] |
| 19 |
Hb_000419_080 |
0.1191395873 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-4 [Jatropha curcas] |
| 20 |
Hb_005747_020 |
0.1197717072 |
- |
- |
bacterial-induced peroxidase [Gossypium schwendimanii] |