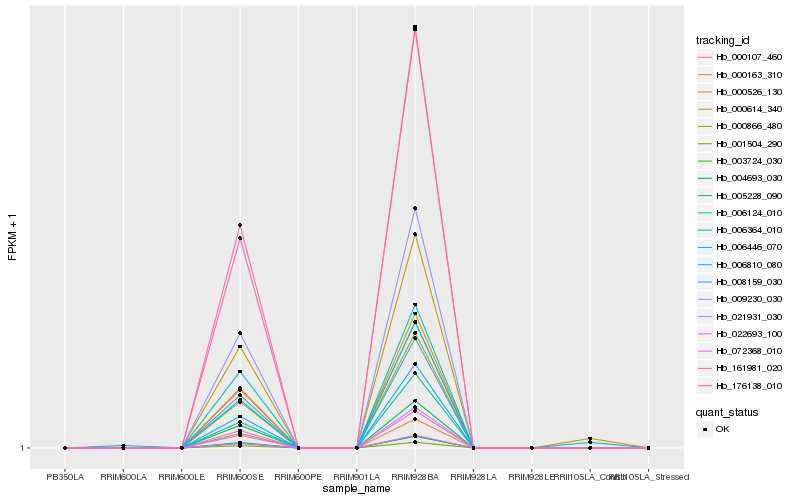
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000614_340 |
0.0 |
- |
- |
Salutaridinol 7-O-acetyltransferase, putative [Ricinus communis] |
| 2 |
Hb_006446_070 |
0.0801898598 |
- |
- |
Zinc-binding dehydrogenase family protein [Theobroma cacao] |
| 3 |
Hb_000526_130 |
0.1023637936 |
- |
- |
PREDICTED: cytochrome P450 705A5-like [Jatropha curcas] |
| 4 |
Hb_009230_030 |
0.1024004743 |
- |
- |
- |
| 5 |
Hb_003724_030 |
0.1024405987 |
- |
- |
- |
| 6 |
Hb_001504_290 |
0.1025229757 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 7 |
Hb_000866_480 |
0.1031110652 |
- |
- |
- |
| 8 |
Hb_000107_460 |
0.1031804553 |
- |
- |
PREDICTED: uncharacterized protein LOC104585930 [Nelumbo nucifera] |
| 9 |
Hb_022693_100 |
0.1032226981 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 10 |
Hb_006810_080 |
0.1039892728 |
- |
- |
hypothetical protein EUGRSUZ_C015472, partial [Eucalyptus grandis] |
| 11 |
Hb_176138_010 |
0.1040320589 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 22 [Jatropha curcas] |
| 12 |
Hb_008159_030 |
0.1055372426 |
- |
- |
hypothetical protein B456_004G088300 [Gossypium raimondii] |
| 13 |
Hb_006124_010 |
0.1069101929 |
- |
- |
- |
| 14 |
Hb_072368_010 |
0.1082936862 |
- |
- |
PREDICTED: uncharacterized protein LOC104800023, partial [Tarenaya hassleriana] |
| 15 |
Hb_161981_020 |
0.1090336792 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 16 |
Hb_004693_030 |
0.1091812107 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Malus domestica] |
| 17 |
Hb_005228_090 |
0.1101474239 |
- |
- |
PREDICTED: magnesium transporter MRS2-3-like [Citrus sinensis] |
| 18 |
Hb_006364_010 |
0.1103863806 |
- |
- |
- |
| 19 |
Hb_021931_030 |
0.110912578 |
- |
- |
hypothetical protein JCGZ_04974 [Jatropha curcas] |
| 20 |
Hb_000163_310 |
0.1141602007 |
- |
- |
PREDICTED: ABC transporter C family member 4-like [Malus domestica] |