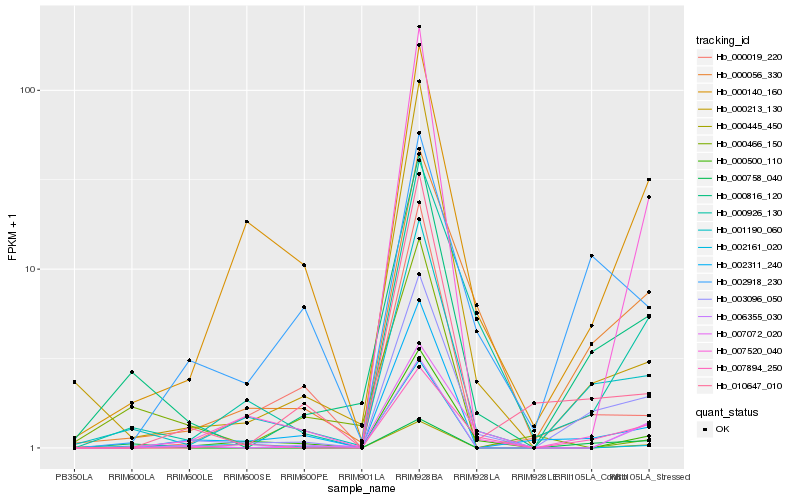
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000816_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649214 [Jatropha curcas] |
| 2 |
Hb_001190_060 |
0.1455422337 |
transcription factor |
TF Family: GRAS |
PREDICTED: nodulation-signaling pathway 1 protein [Jatropha curcas] |
| 3 |
Hb_003096_050 |
0.169175363 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000758_040 |
0.2154462885 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF098 [Jatropha curcas] |
| 5 |
Hb_000056_330 |
0.2171208659 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_000213_130 |
0.2216748991 |
transcription factor |
TF Family: HSF |
Heat Stress Transcription Factor family protein [Populus trichocarpa] |
| 7 |
Hb_010647_010 |
0.2296537023 |
- |
- |
PREDICTED: protein NP24-like [Eucalyptus grandis] |
| 8 |
Hb_002311_240 |
0.2370504271 |
- |
- |
hypothetical protein JCGZ_18937 [Jatropha curcas] |
| 9 |
Hb_007520_040 |
0.2388178718 |
- |
- |
PREDICTED: carbonic anhydrase 2-like [Jatropha curcas] |
| 10 |
Hb_006355_030 |
0.2405936403 |
transcription factor |
TF Family: Orphans |
sensory transduction histidine kinase bacterial, putative [Ricinus communis] |
| 11 |
Hb_000926_130 |
0.2421457343 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 12 |
Hb_007072_020 |
0.2439591693 |
transcription factor |
TF Family: bHLH |
lMYC5 [Hevea brasiliensis] |
| 13 |
Hb_007894_250 |
0.2493214889 |
- |
- |
hypothetical protein JCGZ_06575 [Jatropha curcas] |
| 14 |
Hb_000445_450 |
0.2500174362 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase-like [Jatropha curcas] |
| 15 |
Hb_002161_020 |
0.2591501169 |
transcription factor |
TF Family: bZIP |
hypothetical protein POPTR_0006s05770g, partial [Populus trichocarpa] |
| 16 |
Hb_000140_160 |
0.262313142 |
- |
- |
PREDICTED: neurofilament light polypeptide-like [Gossypium raimondii] |
| 17 |
Hb_000500_110 |
0.2624730353 |
- |
- |
Blue copper protein precursor, putative [Ricinus communis] |
| 18 |
Hb_000466_150 |
0.2659236882 |
- |
- |
hypothetical protein JCGZ_02123 [Jatropha curcas] |
| 19 |
Hb_000019_220 |
0.2662455379 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF113-like [Jatropha curcas] |
| 20 |
Hb_002918_230 |
0.271182768 |
- |
- |
PREDICTED: probable glutathione S-transferase [Gossypium raimondii] |