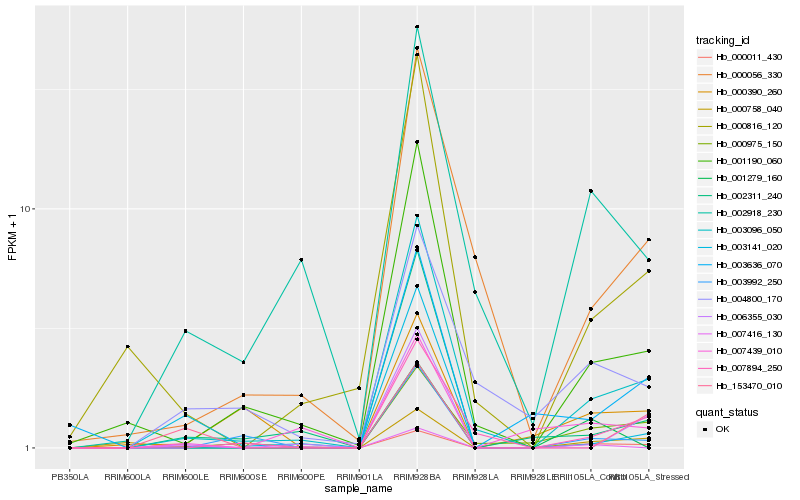
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000011_430 |
0.0 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 2 |
Hb_003096_050 |
0.221957859 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000975_150 |
0.2433959083 |
- |
- |
PREDICTED: phosphatidylinositol/phosphatidylcholine transfer protein SFH8 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000758_040 |
0.2679242012 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF098 [Jatropha curcas] |
| 5 |
Hb_002918_230 |
0.268452414 |
- |
- |
PREDICTED: probable glutathione S-transferase [Gossypium raimondii] |
| 6 |
Hb_000816_120 |
0.2744504945 |
- |
- |
PREDICTED: uncharacterized protein LOC105649214 [Jatropha curcas] |
| 7 |
Hb_001190_060 |
0.2761450548 |
transcription factor |
TF Family: GRAS |
PREDICTED: nodulation-signaling pathway 1 protein [Jatropha curcas] |
| 8 |
Hb_004800_170 |
0.2934475957 |
- |
- |
PREDICTED: cytochrome P450 90B1 [Jatropha curcas] |
| 9 |
Hb_007416_130 |
0.2943988259 |
- |
- |
PREDICTED: UDP-glycosyltransferase 73B4-like [Jatropha curcas] |
| 10 |
Hb_007439_010 |
0.2963457898 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor RAX2-like [Populus euphratica] |
| 11 |
Hb_007894_250 |
0.2989156495 |
- |
- |
hypothetical protein JCGZ_06575 [Jatropha curcas] |
| 12 |
Hb_001279_160 |
0.3022094016 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_003141_020 |
0.3040388205 |
- |
- |
PREDICTED: chlorophyllase-1-like [Jatropha curcas] |
| 14 |
Hb_003636_070 |
0.3052801816 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000390_260 |
0.3056859808 |
transcription factor |
TF Family: MYB |
Myb domain protein 107 [Theobroma cacao] |
| 16 |
Hb_153470_010 |
0.3062335684 |
- |
- |
PREDICTED: uncharacterized protein LOC104804963 [Tarenaya hassleriana] |
| 17 |
Hb_003992_250 |
0.3094786546 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 2-like [Glycine max] |
| 18 |
Hb_000056_330 |
0.3104908586 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 19 |
Hb_002311_240 |
0.3113510138 |
- |
- |
hypothetical protein JCGZ_18937 [Jatropha curcas] |
| 20 |
Hb_006355_030 |
0.3146654405 |
transcription factor |
TF Family: Orphans |
sensory transduction histidine kinase bacterial, putative [Ricinus communis] |