| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
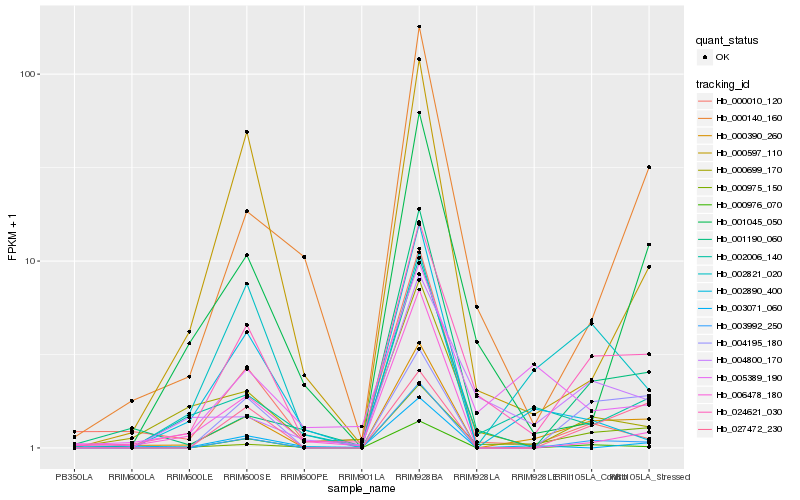
Hb_000390_260 |
0.0 |
transcription factor |
TF Family: MYB |
Myb domain protein 107 [Theobroma cacao] |
| 2 |
Hb_024621_030 |
0.1763025896 |
- |
- |
tetraketide alpha-pyrone reductase 2 [Jatropha curcas] |
| 3 |
Hb_003992_250 |
0.2023286528 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 2-like [Glycine max] |
| 4 |
Hb_000975_150 |
0.2147126917 |
- |
- |
PREDICTED: phosphatidylinositol/phosphatidylcholine transfer protein SFH8 isoform X1 [Jatropha curcas] |
| 5 |
Hb_004195_180 |
0.2192379345 |
- |
- |
unnamed protein product [Coffea canephora] |
| 6 |
Hb_002006_140 |
0.2284777611 |
transcription factor |
TF Family: ERF |
hypothetical protein POPTR_0012s11030g [Populus trichocarpa] |
| 7 |
Hb_027472_230 |
0.2355119264 |
- |
- |
PREDICTED: auxin-induced protein 15A-like [Jatropha curcas] |
| 8 |
Hb_000976_070 |
0.2429275901 |
- |
- |
gulonolactone oxidase, putative [Ricinus communis] |
| 9 |
Hb_001190_060 |
0.2605482905 |
transcription factor |
TF Family: GRAS |
PREDICTED: nodulation-signaling pathway 1 protein [Jatropha curcas] |
| 10 |
Hb_002821_020 |
0.2637811678 |
desease resistance |
Gene Name: ABC_tran |
PREDICTED: ABC transporter C family member 4 [Vitis vinifera] |
| 11 |
Hb_000140_160 |
0.2666175937 |
- |
- |
PREDICTED: neurofilament light polypeptide-like [Gossypium raimondii] |
| 12 |
Hb_004800_170 |
0.2676456234 |
- |
- |
PREDICTED: cytochrome P450 90B1 [Jatropha curcas] |
| 13 |
Hb_000699_170 |
0.2709463905 |
- |
- |
myosin XI, putative [Ricinus communis] |
| 14 |
Hb_002890_400 |
0.2756820696 |
- |
- |
2-nitropropane dioxygenase precursor, putative [Ricinus communis] |
| 15 |
Hb_003071_060 |
0.2757681559 |
- |
- |
PREDICTED: beta-galactosidase 15-like [Jatropha curcas] |
| 16 |
Hb_001045_050 |
0.2791486026 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0001s33560g [Populus trichocarpa] |
| 17 |
Hb_005389_190 |
0.281772077 |
- |
- |
glutamate receptor 2 plant, putative [Ricinus communis] |
| 18 |
Hb_006478_180 |
0.2836088064 |
- |
- |
Nonspecific lipid-transfer protein precursor, putative [Ricinus communis] |
| 19 |
Hb_000597_110 |
0.2845258354 |
- |
- |
PREDICTED: VQ motif-containing protein 22-like [Jatropha curcas] |
| 20 |
Hb_000010_120 |
0.2860466034 |
- |
- |
PREDICTED: cytochrome P450 71A26-like [Jatropha curcas] |