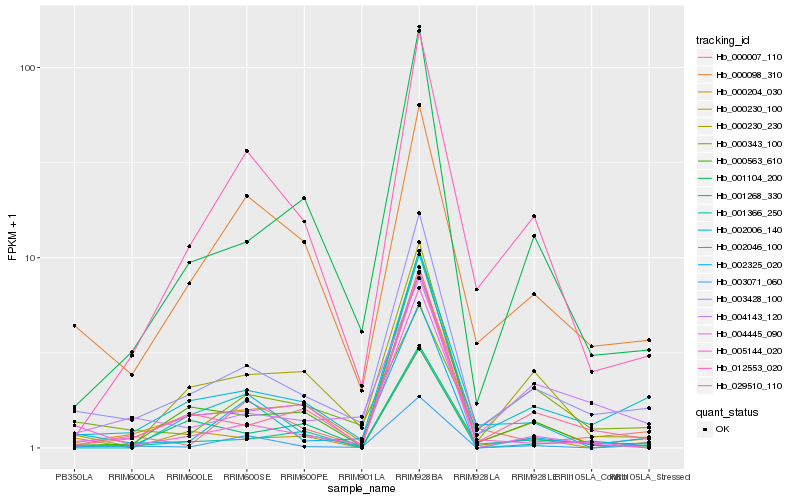
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003428_100 |
0.0 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 2 |
Hb_000204_030 |
0.1287653524 |
- |
- |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 3 |
Hb_029510_110 |
0.1488996347 |
- |
- |
PREDICTED: protein PLANT CADMIUM RESISTANCE 8-like [Jatropha curcas] |
| 4 |
Hb_001104_200 |
0.1528339711 |
- |
- |
Phytosulfokine 3 precursor, putative [Theobroma cacao] |
| 5 |
Hb_002046_100 |
0.1655363927 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB82 [Jatropha curcas] |
| 6 |
Hb_002325_020 |
0.1682656641 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 7 |
Hb_004143_120 |
0.173971587 |
- |
- |
PREDICTED: serine/threonine-protein kinase CTR1 [Jatropha curcas] |
| 8 |
Hb_000230_230 |
0.177350398 |
- |
- |
hydrolase, putative [Ricinus communis] |
| 9 |
Hb_001268_330 |
0.1818016551 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 10 |
Hb_000098_310 |
0.1856235126 |
- |
- |
RecName: Full=Anthocyanidin 3-O-glucosyltransferase 1; AltName: Full=Flavonol 3-O-glucosyltransferase 1; AltName: Full=UDP-glucose flavonoid 3-O-glucosyltransferase 1 [Manihot esculenta] |
| 11 |
Hb_012553_020 |
0.1862537146 |
- |
- |
hypothetical protein JCGZ_09282 [Jatropha curcas] |
| 12 |
Hb_002006_140 |
0.1911476886 |
transcription factor |
TF Family: ERF |
hypothetical protein POPTR_0012s11030g [Populus trichocarpa] |
| 13 |
Hb_000230_100 |
0.1915800544 |
- |
- |
PREDICTED: acyltransferase-like protein At1g54570, chloroplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_000343_100 |
0.1954780636 |
- |
- |
PREDICTED: regulator of telomere elongation helicase 1 homolog [Jatropha curcas] |
| 15 |
Hb_000007_110 |
0.1973003079 |
- |
- |
PREDICTED: peroxidase 11 [Jatropha curcas] |
| 16 |
Hb_000563_610 |
0.1998393683 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase LIN-1 [Jatropha curcas] |
| 17 |
Hb_005144_020 |
0.2033374379 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0009s01270g [Populus trichocarpa] |
| 18 |
Hb_004445_090 |
0.2043615375 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001366_250 |
0.2055859553 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 4-like [Jatropha curcas] |
| 20 |
Hb_003071_060 |
0.2064077315 |
- |
- |
PREDICTED: beta-galactosidase 15-like [Jatropha curcas] |