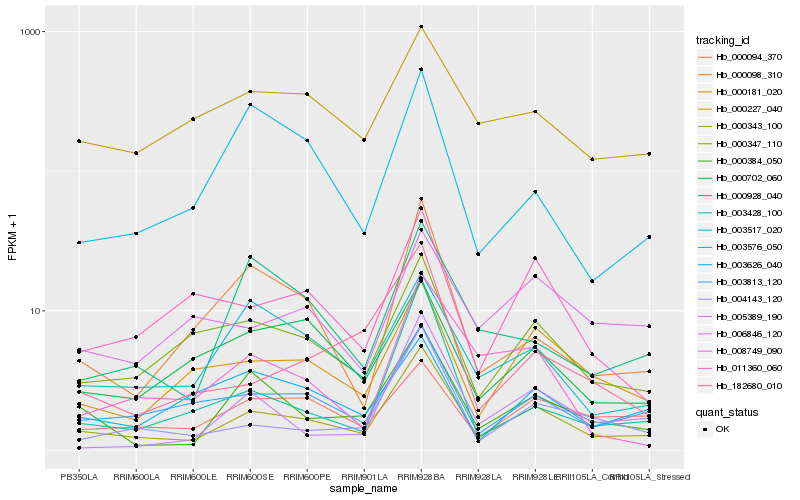
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000343_100 |
0.0 |
- |
- |
PREDICTED: regulator of telomere elongation helicase 1 homolog [Jatropha curcas] |
| 2 |
Hb_003813_120 |
0.1469702138 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 3 |
Hb_000384_050 |
0.1554781174 |
- |
- |
hypothetical protein POPTR_0077s002402g, partial [Populus trichocarpa] |
| 4 |
Hb_004143_120 |
0.1643340764 |
- |
- |
PREDICTED: serine/threonine-protein kinase CTR1 [Jatropha curcas] |
| 5 |
Hb_000181_020 |
0.1648981042 |
- |
- |
PREDICTED: D-amino-acid transaminase, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_000347_110 |
0.1653300161 |
- |
- |
BnaA05g25330D [Brassica napus] |
| 7 |
Hb_000098_310 |
0.187184989 |
- |
- |
RecName: Full=Anthocyanidin 3-O-glucosyltransferase 1; AltName: Full=Flavonol 3-O-glucosyltransferase 1; AltName: Full=UDP-glucose flavonoid 3-O-glucosyltransferase 1 [Manihot esculenta] |
| 8 |
Hb_011360_060 |
0.1903885505 |
- |
- |
glycogen phosphorylase, putative [Ricinus communis] |
| 9 |
Hb_003517_020 |
0.1911342625 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter C family member 3-like [Jatropha curcas] |
| 10 |
Hb_182680_010 |
0.1951137958 |
- |
- |
hypothetical protein JCGZ_07831 [Jatropha curcas] |
| 11 |
Hb_003428_100 |
0.1954780636 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 12 |
Hb_000094_370 |
0.1968350484 |
- |
- |
- |
| 13 |
Hb_005389_190 |
0.1980445404 |
- |
- |
glutamate receptor 2 plant, putative [Ricinus communis] |
| 14 |
Hb_000227_040 |
0.1999457534 |
- |
- |
Stem-specific protein TSJT1, putative [Ricinus communis] |
| 15 |
Hb_000702_060 |
0.2032448603 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_008749_090 |
0.2059167214 |
- |
- |
hypothetical protein CICLE_v10017161mg [Citrus clementina] |
| 17 |
Hb_000928_040 |
0.2073188408 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 18 |
Hb_003576_050 |
0.2108261563 |
transcription factor |
TF Family: EIL |
ETHYLENE-INSENSITIVE3 protein, putative [Ricinus communis] |
| 19 |
Hb_006846_120 |
0.2121376148 |
- |
- |
Succinate/fumarate mitochondrial transporter, putative [Ricinus communis] |
| 20 |
Hb_003626_040 |
0.2122517058 |
- |
- |
PREDICTED: probable 2-oxoglutarate-dependent dioxygenase At3g49630 isoform X2 [Jatropha curcas] |