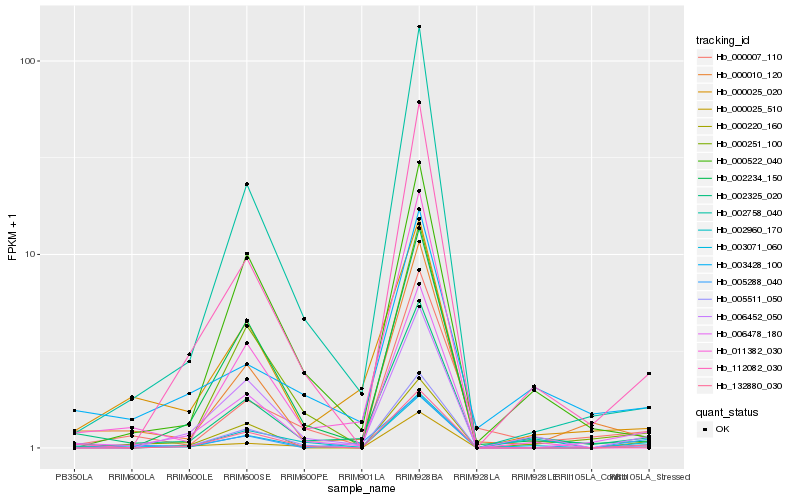
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003071_060 |
0.0 |
- |
- |
PREDICTED: beta-galactosidase 15-like [Jatropha curcas] |
| 2 |
Hb_002325_020 |
0.1208793168 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 3 |
Hb_006478_180 |
0.1700604996 |
- |
- |
Nonspecific lipid-transfer protein precursor, putative [Ricinus communis] |
| 4 |
Hb_006452_050 |
0.1802298178 |
- |
- |
PREDICTED: GABA transporter 1-like [Jatropha curcas] |
| 5 |
Hb_112082_030 |
0.187634712 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 6 |
Hb_002960_170 |
0.1935638246 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At5g57670 [Jatropha curcas] |
| 7 |
Hb_132880_030 |
0.1953921984 |
- |
- |
hypothetical protein JCGZ_10325 [Jatropha curcas] |
| 8 |
Hb_000010_120 |
0.1956871279 |
- |
- |
PREDICTED: cytochrome P450 71A26-like [Jatropha curcas] |
| 9 |
Hb_000025_020 |
0.20152282 |
- |
- |
PREDICTED: cytochrome P450 71B34-like [Jatropha curcas] |
| 10 |
Hb_000007_110 |
0.2057576281 |
- |
- |
PREDICTED: peroxidase 11 [Jatropha curcas] |
| 11 |
Hb_003428_100 |
0.2064077315 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 12 |
Hb_000251_100 |
0.208573039 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 13 |
Hb_002758_040 |
0.2096806893 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 14 |
Hb_011382_030 |
0.211406154 |
- |
- |
Dehydration-responsive protein RD22 precursor, putative [Ricinus communis] |
| 15 |
Hb_005511_050 |
0.2120040238 |
- |
- |
hypothetical protein POPTR_0008s04130g [Populus trichocarpa] |
| 16 |
Hb_000220_160 |
0.2130986988 |
- |
- |
hypothetical protein B456_003G184800 [Gossypium raimondii] |
| 17 |
Hb_002234_150 |
0.2143886508 |
- |
- |
(S)-N-methylcoclaurine 3'-hydroxylase isozyme, putative [Ricinus communis] |
| 18 |
Hb_000025_510 |
0.2158326977 |
- |
- |
PREDICTED: receptor-like protein 12 [Vitis vinifera] |
| 19 |
Hb_005288_040 |
0.2170603165 |
- |
- |
DUF26 domain-containing protein 2 precursor, putative [Ricinus communis] |
| 20 |
Hb_000522_040 |
0.2171619534 |
- |
- |
Desacetoxyvindoline 4-hydroxylase, putative [Ricinus communis] |