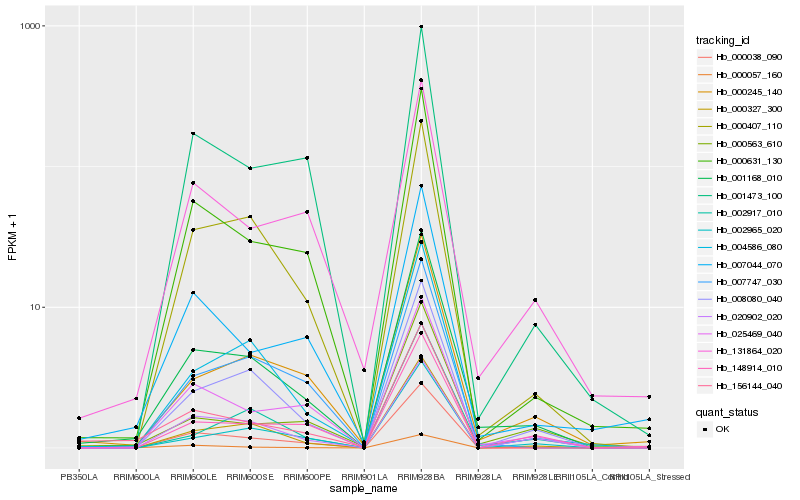
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001168_010 |
0.0 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 1 [Jatropha curcas] |
| 2 |
Hb_000327_300 |
0.104699688 |
- |
- |
PREDICTED: ABC transporter G family member 6-like [Jatropha curcas] |
| 3 |
Hb_000245_140 |
0.1116717986 |
- |
- |
PREDICTED: CASP-like protein 3A1 [Jatropha curcas] |
| 4 |
Hb_000631_130 |
0.1126026182 |
- |
- |
PREDICTED: flavonoid 3'-monooxygenase [Jatropha curcas] |
| 5 |
Hb_004586_080 |
0.1177506819 |
- |
- |
hypothetical protein POPTR_0002s10520g [Populus trichocarpa] |
| 6 |
Hb_008080_040 |
0.1184077056 |
- |
- |
hypothetical protein EUGRSUZ_D00834 [Eucalyptus grandis] |
| 7 |
Hb_000038_090 |
0.1185229284 |
transcription factor |
TF Family: NAC |
hypothetical protein POPTR_0002s05820g [Populus trichocarpa] |
| 8 |
Hb_025469_040 |
0.121428123 |
- |
- |
PREDICTED: acidic mammalian chitinase-like [Jatropha curcas] |
| 9 |
Hb_002965_020 |
0.1269388327 |
- |
- |
hypothetical protein JCGZ_18947 [Jatropha curcas] |
| 10 |
Hb_000407_110 |
0.1283494482 |
- |
- |
ripening-induced ACC oxidase [Carica papaya] |
| 11 |
Hb_007044_070 |
0.1329055796 |
- |
- |
PREDICTED: tetraketide alpha-pyrone reductase 1-like [Jatropha curcas] |
| 12 |
Hb_156144_040 |
0.1337512964 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 13 |
Hb_000563_610 |
0.1344828456 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase LIN-1 [Jatropha curcas] |
| 14 |
Hb_007747_030 |
0.1403881347 |
- |
- |
PREDICTED: WD repeat-containing protein LWD2-like [Jatropha curcas] |
| 15 |
Hb_148914_010 |
0.1412784636 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 16 |
Hb_002917_010 |
0.1416474502 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1 [Jatropha curcas] |
| 17 |
Hb_020902_020 |
0.1446023506 |
- |
- |
hypothetical protein JCGZ_07820 [Jatropha curcas] |
| 18 |
Hb_131864_020 |
0.1448675612 |
- |
- |
PREDICTED: anthocyanidin reductase [Jatropha curcas] |
| 19 |
Hb_001473_100 |
0.1457524812 |
- |
- |
chalcone synthase 1 [Jatropha curcas] |
| 20 |
Hb_000057_160 |
0.1472502365 |
- |
- |
hypothetical protein JCGZ_06279 [Jatropha curcas] |