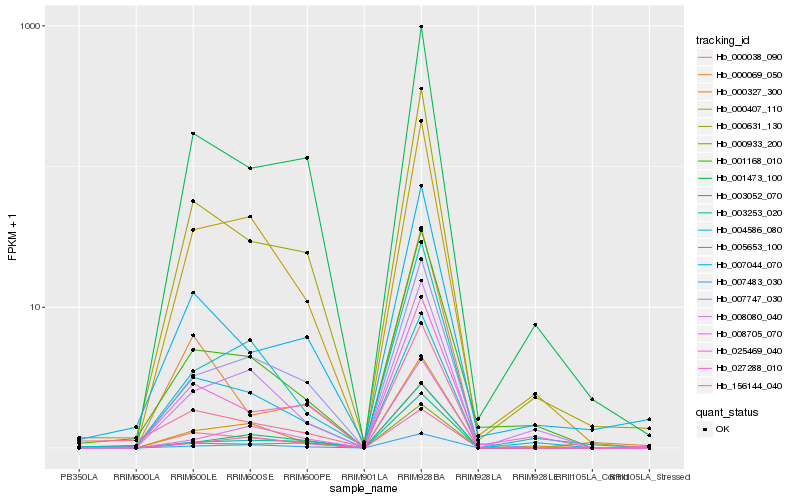
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000038_090 |
0.0 |
transcription factor |
TF Family: NAC |
hypothetical protein POPTR_0002s05820g [Populus trichocarpa] |
| 2 |
Hb_000631_130 |
0.0660923963 |
- |
- |
PREDICTED: flavonoid 3'-monooxygenase [Jatropha curcas] |
| 3 |
Hb_001473_100 |
0.1075379661 |
- |
- |
chalcone synthase 1 [Jatropha curcas] |
| 4 |
Hb_000933_200 |
0.1097854494 |
- |
- |
Cationic peroxidase 2 precursor, putative [Ricinus communis] |
| 5 |
Hb_000327_300 |
0.1109620117 |
- |
- |
PREDICTED: ABC transporter G family member 6-like [Jatropha curcas] |
| 6 |
Hb_007747_030 |
0.112469419 |
- |
- |
PREDICTED: WD repeat-containing protein LWD2-like [Jatropha curcas] |
| 7 |
Hb_000407_110 |
0.1129020094 |
- |
- |
ripening-induced ACC oxidase [Carica papaya] |
| 8 |
Hb_007483_030 |
0.1137521483 |
- |
- |
- |
| 9 |
Hb_001168_010 |
0.1185229284 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 1 [Jatropha curcas] |
| 10 |
Hb_003253_020 |
0.1226733492 |
transcription factor |
TF Family: B3 |
transcription factor, putative [Ricinus communis] |
| 11 |
Hb_025469_040 |
0.1256104468 |
- |
- |
PREDICTED: acidic mammalian chitinase-like [Jatropha curcas] |
| 12 |
Hb_156144_040 |
0.1272329084 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 13 |
Hb_004586_080 |
0.1278693457 |
- |
- |
hypothetical protein POPTR_0002s10520g [Populus trichocarpa] |
| 14 |
Hb_000069_050 |
0.1299667022 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 308-like [Populus euphratica] |
| 15 |
Hb_007044_070 |
0.130327103 |
- |
- |
PREDICTED: tetraketide alpha-pyrone reductase 1-like [Jatropha curcas] |
| 16 |
Hb_005653_100 |
0.1306215505 |
- |
- |
hypothetical protein POPTR_0004s19580g [Populus trichocarpa] |
| 17 |
Hb_008080_040 |
0.1380650863 |
- |
- |
hypothetical protein EUGRSUZ_D00834 [Eucalyptus grandis] |
| 18 |
Hb_027288_010 |
0.1388690697 |
- |
- |
mutt domain protein, putative [Ricinus communis] |
| 19 |
Hb_008705_070 |
0.1405411329 |
- |
- |
PREDICTED: uncharacterized protein LOC105638774 [Jatropha curcas] |
| 20 |
Hb_003052_070 |
0.1419402809 |
- |
- |
PREDICTED: peroxidase 11 [Jatropha curcas] |