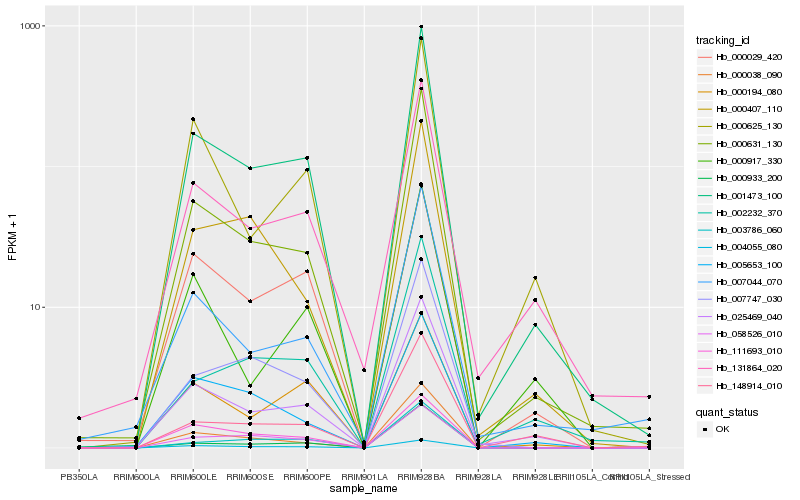
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001473_100 |
0.0 |
- |
- |
chalcone synthase 1 [Jatropha curcas] |
| 2 |
Hb_000631_130 |
0.0669362836 |
- |
- |
PREDICTED: flavonoid 3'-monooxygenase [Jatropha curcas] |
| 3 |
Hb_025469_040 |
0.0722138315 |
- |
- |
PREDICTED: acidic mammalian chitinase-like [Jatropha curcas] |
| 4 |
Hb_131864_020 |
0.093497338 |
- |
- |
PREDICTED: anthocyanidin reductase [Jatropha curcas] |
| 5 |
Hb_004055_080 |
0.1016206649 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 6 |
Hb_000038_090 |
0.1075379661 |
transcription factor |
TF Family: NAC |
hypothetical protein POPTR_0002s05820g [Populus trichocarpa] |
| 7 |
Hb_007044_070 |
0.1120246979 |
- |
- |
PREDICTED: tetraketide alpha-pyrone reductase 1-like [Jatropha curcas] |
| 8 |
Hb_007747_030 |
0.11375417 |
- |
- |
PREDICTED: WD repeat-containing protein LWD2-like [Jatropha curcas] |
| 9 |
Hb_000625_130 |
0.1168131997 |
- |
- |
PREDICTED: naringenin,2-oxoglutarate 3-dioxygenase [Jatropha curcas] |
| 10 |
Hb_003786_060 |
0.1175913042 |
- |
- |
PREDICTED: transmembrane protein 220 [Jatropha curcas] |
| 11 |
Hb_058526_010 |
0.1179145503 |
- |
- |
- |
| 12 |
Hb_000194_080 |
0.1198854555 |
- |
- |
hypothetical protein JCGZ_18743 [Jatropha curcas] |
| 13 |
Hb_000933_200 |
0.1211422126 |
- |
- |
Cationic peroxidase 2 precursor, putative [Ricinus communis] |
| 14 |
Hb_000029_420 |
0.1263879981 |
- |
- |
PREDICTED: putative lipid-transfer protein DIR1 [Jatropha curcas] |
| 15 |
Hb_148914_010 |
0.127271847 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 16 |
Hb_005653_100 |
0.1278513781 |
- |
- |
hypothetical protein POPTR_0004s19580g [Populus trichocarpa] |
| 17 |
Hb_002232_370 |
0.1302452271 |
- |
- |
PREDICTED: receptor-like protein kinase HAIKU2 [Jatropha curcas] |
| 18 |
Hb_111693_010 |
0.1306674582 |
- |
- |
hypothetical protein JCGZ_21650 [Jatropha curcas] |
| 19 |
Hb_000407_110 |
0.133577311 |
- |
- |
ripening-induced ACC oxidase [Carica papaya] |
| 20 |
Hb_000917_330 |
0.1347099445 |
- |
- |
PREDICTED: leucoanthocyanidin reductase-like [Jatropha curcas] |