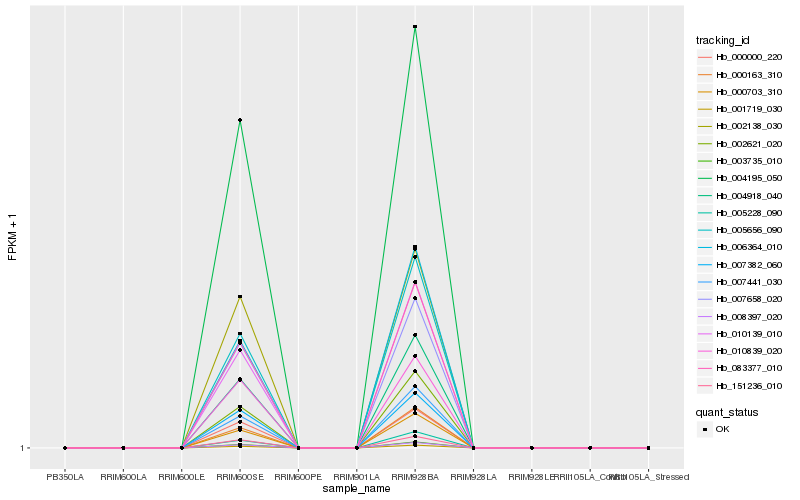
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004918_040 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_003735_010 |
0.0021286265 |
- |
- |
- |
| 3 |
Hb_008397_020 |
0.0022669405 |
- |
- |
polyprotein [Oryza australiensis] |
| 4 |
Hb_001719_030 |
0.0029780897 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 5 |
Hb_083377_010 |
0.0036327323 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_005656_090 |
0.0142103835 |
transcription factor |
TF Family: NAC |
hypothetical protein CICLE_v10008885mg [Citrus clementina] |
| 7 |
Hb_010139_010 |
0.0151939227 |
- |
- |
PREDICTED: uncharacterized protein LOC105645488 [Jatropha curcas] |
| 8 |
Hb_002621_020 |
0.0221840962 |
- |
- |
- |
| 9 |
Hb_000703_310 |
0.0255759285 |
- |
- |
- |
| 10 |
Hb_007658_020 |
0.0278214341 |
- |
- |
Beta-amyrin synthase [Glycine soja] |
| 11 |
Hb_000000_220 |
0.0281495969 |
- |
- |
protein with unknown function [Ricinus communis] |
| 12 |
Hb_007441_030 |
0.0283920358 |
- |
- |
PREDICTED: uclacyanin-2-like [Jatropha curcas] |
| 13 |
Hb_007382_060 |
0.0317320037 |
- |
- |
- |
| 14 |
Hb_004195_050 |
0.0317719604 |
- |
- |
- |
| 15 |
Hb_000163_310 |
0.0336152434 |
- |
- |
PREDICTED: ABC transporter C family member 4-like [Malus domestica] |
| 16 |
Hb_151236_010 |
0.0402307988 |
- |
- |
hypothetical protein POPTR_0010s11560g [Populus trichocarpa] |
| 17 |
Hb_006364_010 |
0.0428730712 |
- |
- |
- |
| 18 |
Hb_005228_090 |
0.043517483 |
- |
- |
PREDICTED: magnesium transporter MRS2-3-like [Citrus sinensis] |
| 19 |
Hb_002138_030 |
0.0438296646 |
- |
- |
- |
| 20 |
Hb_010839_020 |
0.0461703891 |
- |
- |
- |