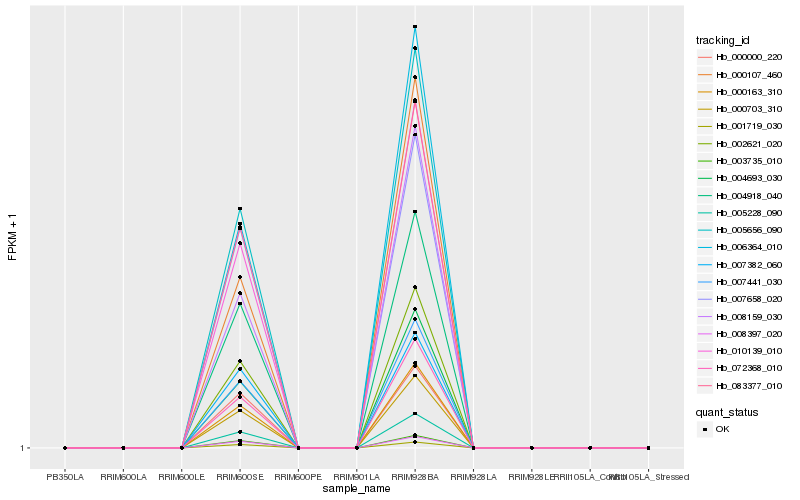
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002621_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_000703_310 |
0.0033926383 |
- |
- |
- |
| 3 |
Hb_007441_030 |
0.0062095816 |
- |
- |
PREDICTED: uclacyanin-2-like [Jatropha curcas] |
| 4 |
Hb_010139_010 |
0.0069911509 |
- |
- |
PREDICTED: uncharacterized protein LOC105645488 [Jatropha curcas] |
| 5 |
Hb_005656_090 |
0.0079747545 |
transcription factor |
TF Family: NAC |
hypothetical protein CICLE_v10008885mg [Citrus clementina] |
| 6 |
Hb_000163_310 |
0.0114347447 |
- |
- |
PREDICTED: ABC transporter C family member 4-like [Malus domestica] |
| 7 |
Hb_006364_010 |
0.0206973533 |
- |
- |
- |
| 8 |
Hb_005228_090 |
0.0213421614 |
- |
- |
PREDICTED: magnesium transporter MRS2-3-like [Citrus sinensis] |
| 9 |
Hb_004918_040 |
0.0221840962 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_004693_030 |
0.0240442507 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Malus domestica] |
| 11 |
Hb_003735_010 |
0.0243122538 |
- |
- |
- |
| 12 |
Hb_008397_020 |
0.0244505344 |
- |
- |
polyprotein [Oryza australiensis] |
| 13 |
Hb_001719_030 |
0.0251615074 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 14 |
Hb_083377_010 |
0.0258159798 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_072368_010 |
0.0266824408 |
- |
- |
PREDICTED: uncharacterized protein LOC104800023, partial [Tarenaya hassleriana] |
| 16 |
Hb_008159_030 |
0.0363408806 |
- |
- |
hypothetical protein B456_004G088300 [Gossypium raimondii] |
| 17 |
Hb_000107_460 |
0.0489479014 |
- |
- |
PREDICTED: uncharacterized protein LOC104585930 [Nelumbo nucifera] |
| 18 |
Hb_007658_020 |
0.0499931963 |
- |
- |
Beta-amyrin synthase [Glycine soja] |
| 19 |
Hb_000000_220 |
0.050321135 |
- |
- |
protein with unknown function [Ricinus communis] |
| 20 |
Hb_007382_060 |
0.0539009804 |
- |
- |
- |