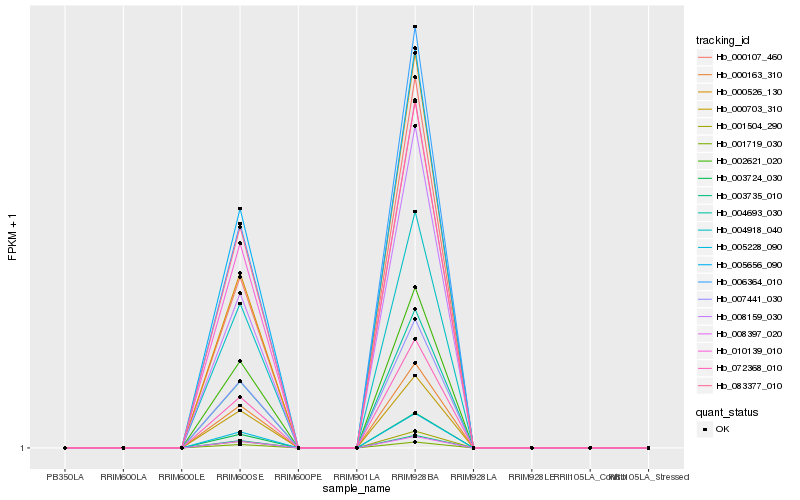
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007441_030 |
0.0 |
- |
- |
PREDICTED: uclacyanin-2-like [Jatropha curcas] |
| 2 |
Hb_000703_310 |
0.0028169688 |
- |
- |
- |
| 3 |
Hb_000163_310 |
0.005225324 |
- |
- |
PREDICTED: ABC transporter C family member 4-like [Malus domestica] |
| 4 |
Hb_002621_020 |
0.0062095816 |
- |
- |
- |
| 5 |
Hb_010139_010 |
0.0132004884 |
- |
- |
PREDICTED: uncharacterized protein LOC105645488 [Jatropha curcas] |
| 6 |
Hb_005656_090 |
0.0141840372 |
transcription factor |
TF Family: NAC |
hypothetical protein CICLE_v10008885mg [Citrus clementina] |
| 7 |
Hb_006364_010 |
0.0144885869 |
- |
- |
- |
| 8 |
Hb_005228_090 |
0.0151334583 |
- |
- |
PREDICTED: magnesium transporter MRS2-3-like [Citrus sinensis] |
| 9 |
Hb_004693_030 |
0.0178358388 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Malus domestica] |
| 10 |
Hb_072368_010 |
0.0204743534 |
- |
- |
PREDICTED: uncharacterized protein LOC104800023, partial [Tarenaya hassleriana] |
| 11 |
Hb_004918_040 |
0.0283920358 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_008159_030 |
0.0301343245 |
- |
- |
hypothetical protein B456_004G088300 [Gossypium raimondii] |
| 13 |
Hb_003735_010 |
0.0305199049 |
- |
- |
- |
| 14 |
Hb_008397_020 |
0.030658166 |
- |
- |
polyprotein [Oryza australiensis] |
| 15 |
Hb_001719_030 |
0.0313690371 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 16 |
Hb_083377_010 |
0.0320234135 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_000107_460 |
0.0427441781 |
- |
- |
PREDICTED: uncharacterized protein LOC104585930 [Nelumbo nucifera] |
| 18 |
Hb_001504_290 |
0.0495921464 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 19 |
Hb_003724_030 |
0.051059161 |
- |
- |
- |
| 20 |
Hb_000526_130 |
0.052992163 |
- |
- |
PREDICTED: cytochrome P450 705A5-like [Jatropha curcas] |