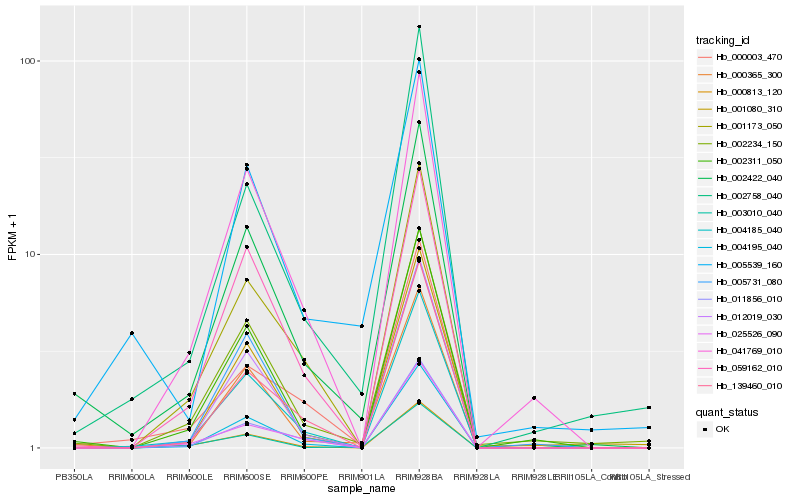
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002422_040 |
0.0 |
- |
- |
hypothetical protein POPTR_0016s02350g [Populus trichocarpa] |
| 2 |
Hb_000365_300 |
0.1030159376 |
- |
- |
PREDICTED: cytochrome P450 714C2-like [Jatropha curcas] |
| 3 |
Hb_004185_040 |
0.1037465911 |
- |
- |
polygalacturonase, putative [Ricinus communis] |
| 4 |
Hb_002234_150 |
0.1099999523 |
- |
- |
(S)-N-methylcoclaurine 3'-hydroxylase isozyme, putative [Ricinus communis] |
| 5 |
Hb_002311_050 |
0.112108012 |
- |
- |
PREDICTED: uncharacterized protein LOC105122837 isoform X2 [Populus euphratica] |
| 6 |
Hb_004195_040 |
0.1149715408 |
- |
- |
salicylic acid methyl transferase [Capsicum annuum] |
| 7 |
Hb_059162_010 |
0.1172520647 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 8 |
Hb_012019_030 |
0.1211148299 |
- |
- |
hypothetical protein POPTR_0005s23360g [Populus trichocarpa] |
| 9 |
Hb_000813_120 |
0.1249865662 |
- |
- |
PREDICTED: serine carboxypeptidase-like 18 [Tarenaya hassleriana] |
| 10 |
Hb_001173_050 |
0.125664909 |
- |
- |
DUF26 domain-containing protein 2 precursor, putative [Ricinus communis] |
| 11 |
Hb_041769_010 |
0.1272185872 |
- |
- |
hypothetical protein JCGZ_24970 [Jatropha curcas] |
| 12 |
Hb_000003_470 |
0.1287158884 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002758_040 |
0.1315596152 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 14 |
Hb_005731_080 |
0.1316555776 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 15 |
Hb_011856_010 |
0.1320985353 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 16 |
Hb_001080_310 |
0.1336858029 |
- |
- |
PREDICTED: ent-copalyl diphosphate synthase, chloroplastic [Jatropha curcas] |
| 17 |
Hb_025526_090 |
0.1338553285 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 [Jatropha curcas] |
| 18 |
Hb_139460_010 |
0.1338567114 |
- |
- |
PREDICTED: beta-glucosidase 18-like [Jatropha curcas] |
| 19 |
Hb_005539_160 |
0.1340335748 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 2 member B7, mitochondrial-like [Jatropha curcas] |
| 20 |
Hb_003010_040 |
0.1379200389 |
- |
- |
hypothetical protein AALP_AA7G248200 [Arabis alpina] |