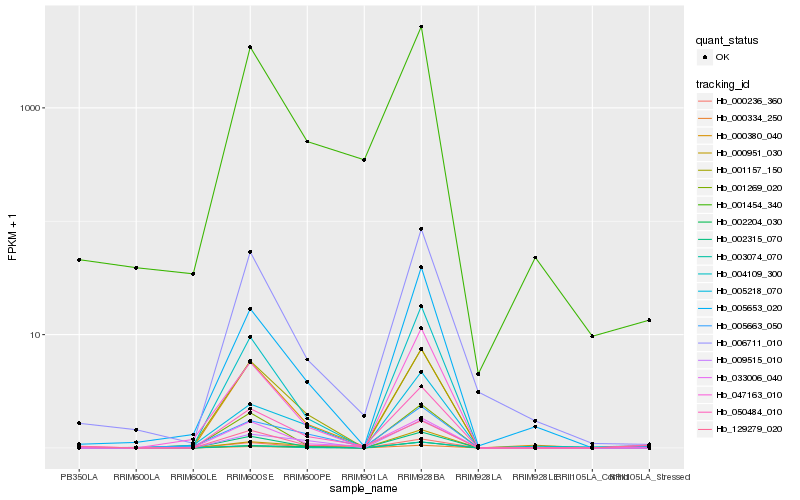
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_050484_010 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_129279_020 |
0.1061306885 |
- |
- |
PREDICTED: putative disease resistance protein At3g14460 [Jatropha curcas] |
| 3 |
Hb_000236_360 |
0.138706897 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase FLS2 [Prunus mume] |
| 4 |
Hb_002315_070 |
0.1414164795 |
- |
- |
PREDICTED: uncharacterized protein LOC105630187 [Jatropha curcas] |
| 5 |
Hb_001454_340 |
0.1444184615 |
- |
- |
ATDI21 [Manihot esculenta] |
| 6 |
Hb_004109_300 |
0.1447483198 |
- |
- |
hypothetical protein POPTR_0003s21800g [Populus trichocarpa] |
| 7 |
Hb_001157_150 |
0.15000718 |
- |
- |
PREDICTED: transmembrane protein 45A-like [Jatropha curcas] |
| 8 |
Hb_005653_020 |
0.1511814491 |
transcription factor |
TF Family: C2H2 |
TRANSPARENT TESTA 1 protein, putative [Ricinus communis] |
| 9 |
Hb_000380_040 |
0.1524156423 |
- |
- |
bel1 homeotic protein, putative [Ricinus communis] |
| 10 |
Hb_001269_020 |
0.1525853776 |
- |
- |
PREDICTED: beta-Amyrin Synthase 2-like [Vitis vinifera] |
| 11 |
Hb_002204_030 |
0.1549683703 |
- |
- |
PREDICTED: uncharacterized protein LOC105764710 isoform X3 [Gossypium raimondii] |
| 12 |
Hb_006711_010 |
0.1572712392 |
- |
- |
cytoplasmic dynein light chain, putative [Ricinus communis] |
| 13 |
Hb_005663_050 |
0.1598166473 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 14 |
Hb_000951_030 |
0.1618726959 |
- |
- |
kinase, putative [Ricinus communis] |
| 15 |
Hb_000334_250 |
0.1625139154 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 16 |
Hb_009515_010 |
0.164180946 |
transcription factor |
TF Family: HB |
hypothetical protein JCGZ_15302 [Jatropha curcas] |
| 17 |
Hb_003074_070 |
0.1645319383 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 18 |
Hb_005218_070 |
0.1660271449 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor RAX3 isoform X2 [Populus euphratica] |
| 19 |
Hb_047163_010 |
0.1666271612 |
- |
- |
PREDICTED: periaxin-like [Populus euphratica] |
| 20 |
Hb_033006_040 |
0.1668708655 |
- |
- |
sulfate transporter, putative [Ricinus communis] |