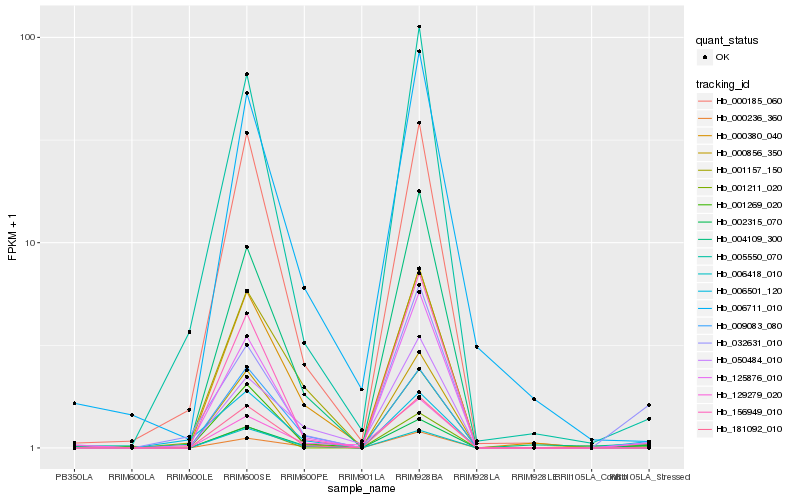
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002315_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105630187 [Jatropha curcas] |
| 2 |
Hb_129279_020 |
0.1204067455 |
- |
- |
PREDICTED: putative disease resistance protein At3g14460 [Jatropha curcas] |
| 3 |
Hb_050484_010 |
0.1414164795 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_001269_020 |
0.1554762343 |
- |
- |
PREDICTED: beta-Amyrin Synthase 2-like [Vitis vinifera] |
| 5 |
Hb_006418_010 |
0.1767730737 |
- |
- |
PREDICTED: cytochrome P450 76A1-like [Jatropha curcas] |
| 6 |
Hb_001157_150 |
0.1826647267 |
- |
- |
PREDICTED: transmembrane protein 45A-like [Jatropha curcas] |
| 7 |
Hb_004109_300 |
0.1856027368 |
- |
- |
hypothetical protein POPTR_0003s21800g [Populus trichocarpa] |
| 8 |
Hb_032631_010 |
0.1875187522 |
- |
- |
PREDICTED: uncharacterized protein LOC105638774 [Jatropha curcas] |
| 9 |
Hb_000236_360 |
0.1882580868 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase FLS2 [Prunus mume] |
| 10 |
Hb_181092_010 |
0.1895322595 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 11 |
Hb_000380_040 |
0.1908151299 |
- |
- |
bel1 homeotic protein, putative [Ricinus communis] |
| 12 |
Hb_000185_060 |
0.1972542875 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic [Jatropha curcas] |
| 13 |
Hb_009083_080 |
0.1986975202 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_006711_010 |
0.19917699 |
- |
- |
cytoplasmic dynein light chain, putative [Ricinus communis] |
| 15 |
Hb_156949_010 |
0.2018410464 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |
| 16 |
Hb_125876_010 |
0.20295444 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_006501_120 |
0.2032034986 |
- |
- |
hypothetical protein JCGZ_07378 [Jatropha curcas] |
| 18 |
Hb_000856_350 |
0.2046106044 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor 36 [Jatropha curcas] |
| 19 |
Hb_005550_070 |
0.2052370468 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 75 [Populus euphratica] |
| 20 |
Hb_001211_020 |
0.2065766003 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0002s11440g [Populus trichocarpa] |