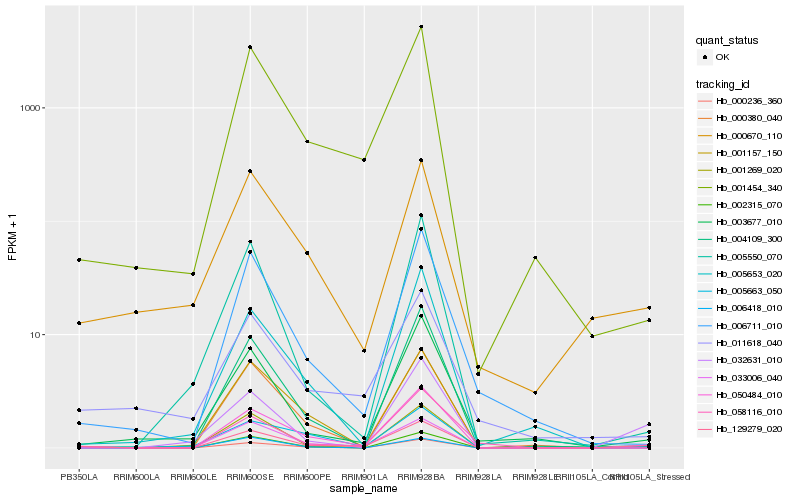
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_129279_020 |
0.0 |
- |
- |
PREDICTED: putative disease resistance protein At3g14460 [Jatropha curcas] |
| 2 |
Hb_050484_010 |
0.1061306885 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 3 |
Hb_002315_070 |
0.1204067455 |
- |
- |
PREDICTED: uncharacterized protein LOC105630187 [Jatropha curcas] |
| 4 |
Hb_001454_340 |
0.1698107936 |
- |
- |
ATDI21 [Manihot esculenta] |
| 5 |
Hb_001269_020 |
0.1816181525 |
- |
- |
PREDICTED: beta-Amyrin Synthase 2-like [Vitis vinifera] |
| 6 |
Hb_033006_040 |
0.1971260597 |
- |
- |
sulfate transporter, putative [Ricinus communis] |
| 7 |
Hb_006711_010 |
0.1992657612 |
- |
- |
cytoplasmic dynein light chain, putative [Ricinus communis] |
| 8 |
Hb_003677_010 |
0.2071433592 |
transcription factor |
TF Family: SRS |
PREDICTED: protein SHI RELATED SEQUENCE 1 [Jatropha curcas] |
| 9 |
Hb_032631_010 |
0.2073668674 |
- |
- |
PREDICTED: uncharacterized protein LOC105638774 [Jatropha curcas] |
| 10 |
Hb_004109_300 |
0.2080867198 |
- |
- |
hypothetical protein POPTR_0003s21800g [Populus trichocarpa] |
| 11 |
Hb_000380_040 |
0.2085809323 |
- |
- |
bel1 homeotic protein, putative [Ricinus communis] |
| 12 |
Hb_000236_360 |
0.212476708 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase FLS2 [Prunus mume] |
| 13 |
Hb_001157_150 |
0.2143097416 |
- |
- |
PREDICTED: transmembrane protein 45A-like [Jatropha curcas] |
| 14 |
Hb_058116_010 |
0.2150246258 |
- |
- |
Cycloartenol synthase, putative [Ricinus communis] |
| 15 |
Hb_000670_110 |
0.2153639482 |
- |
- |
l-asparaginase, putative [Ricinus communis] |
| 16 |
Hb_011618_040 |
0.2175227594 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_005653_020 |
0.2237843206 |
transcription factor |
TF Family: C2H2 |
TRANSPARENT TESTA 1 protein, putative [Ricinus communis] |
| 18 |
Hb_005550_070 |
0.2243247717 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 75 [Populus euphratica] |
| 19 |
Hb_005663_050 |
0.2243382327 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 20 |
Hb_006418_010 |
0.2262514188 |
- |
- |
PREDICTED: cytochrome P450 76A1-like [Jatropha curcas] |