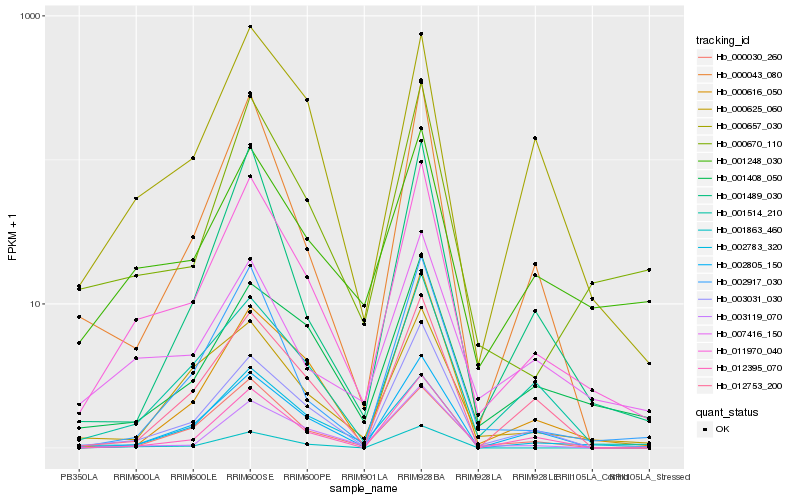
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011970_040 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_003031_030 |
0.1276935333 |
- |
- |
hypothetical protein JCGZ_21288 [Jatropha curcas] |
| 3 |
Hb_000670_110 |
0.136253668 |
- |
- |
l-asparaginase, putative [Ricinus communis] |
| 4 |
Hb_001514_210 |
0.139170345 |
- |
- |
hypothetical protein VITISV_012133 [Vitis vinifera] |
| 5 |
Hb_002783_320 |
0.1431345322 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor E2FC isoform X1 [Jatropha curcas] |
| 6 |
Hb_001863_460 |
0.1453569723 |
transcription factor |
TF Family: MYB |
PREDICTED: protein ODORANT1 [Jatropha curcas] |
| 7 |
Hb_001248_030 |
0.1463426255 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 8 |
Hb_002805_150 |
0.14726084 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 9 |
Hb_003119_070 |
0.1480651175 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 10 |
Hb_002917_030 |
0.1483566901 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase HSL2 [Setaria italica] |
| 11 |
Hb_000043_080 |
0.1485453823 |
- |
- |
lipid binding protein, putative [Ricinus communis] |
| 12 |
Hb_012395_070 |
0.1506140782 |
- |
- |
PREDICTED: probable metal-nicotianamine transporter YSL6 [Jatropha curcas] |
| 13 |
Hb_000657_030 |
0.1532586 |
transcription factor |
TF Family: bZIP |
Ocs element-binding factor, putative [Ricinus communis] |
| 14 |
Hb_000616_050 |
0.1574096624 |
- |
- |
PREDICTED: uncharacterized protein LOC105639906 [Jatropha curcas] |
| 15 |
Hb_007416_150 |
0.1576550719 |
- |
- |
sarcosine oxidase, putative [Ricinus communis] |
| 16 |
Hb_012753_200 |
0.1582690943 |
- |
- |
hypothetical protein JCGZ_20651 [Jatropha curcas] |
| 17 |
Hb_001408_050 |
0.158891453 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001489_030 |
0.1589987958 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 4.6 [Jatropha curcas] |
| 19 |
Hb_000030_260 |
0.1592014684 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat containing protein, putative [Ricinus communis] |
| 20 |
Hb_000625_060 |
0.1617296057 |
- |
- |
Protein P21, putative [Ricinus communis] |