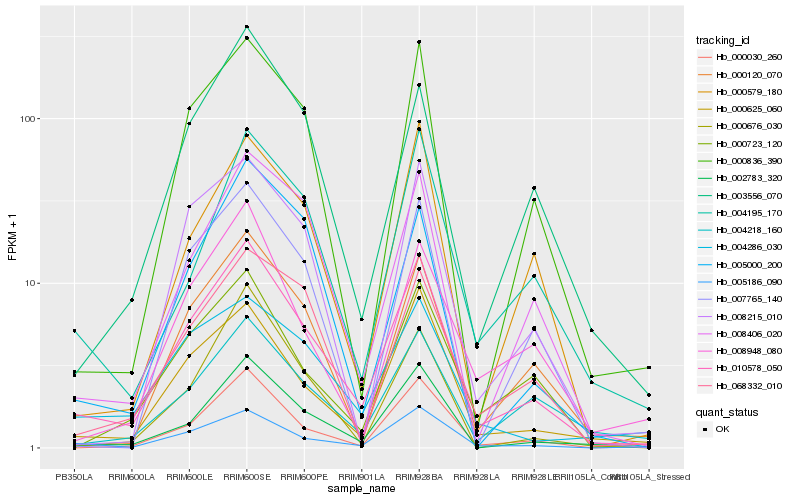
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010578_050 |
0.0 |
transcription factor |
TF Family: bHLH |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_005186_090 |
0.112636113 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 3 |
Hb_002783_320 |
0.1153625844 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor E2FC isoform X1 [Jatropha curcas] |
| 4 |
Hb_000625_060 |
0.119864152 |
- |
- |
Protein P21, putative [Ricinus communis] |
| 5 |
Hb_000030_260 |
0.1290215268 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat containing protein, putative [Ricinus communis] |
| 6 |
Hb_000836_390 |
0.1360517693 |
- |
- |
PREDICTED: neurofilament light polypeptide-like [Gossypium raimondii] |
| 7 |
Hb_000120_070 |
0.1396492663 |
- |
- |
PREDICTED: uncharacterized protein LOC105630351 [Jatropha curcas] |
| 8 |
Hb_008406_020 |
0.141806238 |
- |
- |
PREDICTED: probable aminotransferase TAT2 [Jatropha curcas] |
| 9 |
Hb_000723_120 |
0.1458610002 |
- |
- |
PREDICTED: UDP-glycosyltransferase 86A1 [Jatropha curcas] |
| 10 |
Hb_004218_160 |
0.1515662865 |
- |
- |
NBS-LRR type disease resistance protein [Medicago truncatula] |
| 11 |
Hb_003556_070 |
0.1517129926 |
- |
- |
metal ion binding protein, putative [Ricinus communis] |
| 12 |
Hb_004195_170 |
0.1518357708 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 13 |
Hb_068332_010 |
0.1527058993 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 14 |
Hb_008215_010 |
0.1534954357 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000579_180 |
0.1564681549 |
transcription factor |
TF Family: bZIP |
PREDICTED: ocs element-binding factor 1-like [Jatropha curcas] |
| 16 |
Hb_004286_030 |
0.156883299 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor ANT [Jatropha curcas] |
| 17 |
Hb_008948_080 |
0.1614399762 |
- |
- |
PREDICTED: TBC1 domain family member 13-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000676_030 |
0.1634524881 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-B [Jatropha curcas] |
| 19 |
Hb_007765_140 |
0.1641330991 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 8.1-like [Jatropha curcas] |
| 20 |
Hb_005000_200 |
0.1646757093 |
- |
- |
PREDICTED: uncharacterized protein LOC105637887 [Jatropha curcas] |