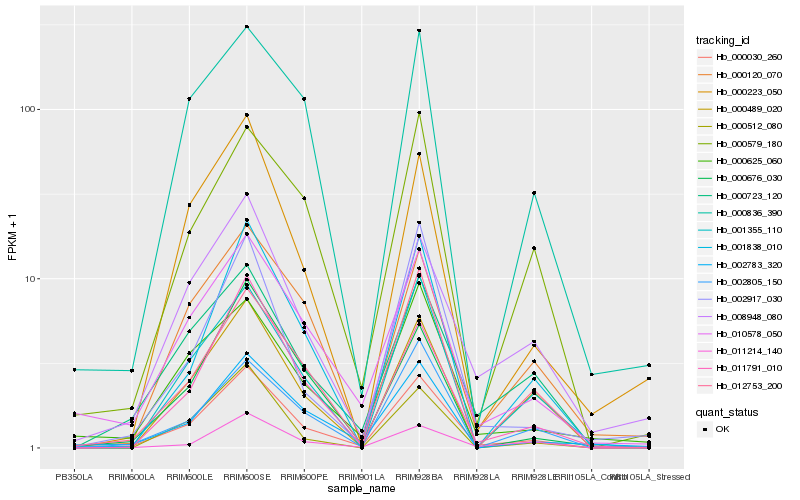
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000030_260 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat containing protein, putative [Ricinus communis] |
| 2 |
Hb_011791_010 |
0.1276417305 |
- |
- |
PREDICTED: cytochrome P450 83B1-like [Jatropha curcas] |
| 3 |
Hb_010578_050 |
0.1290215268 |
transcription factor |
TF Family: bHLH |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002783_320 |
0.1345028876 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor E2FC isoform X1 [Jatropha curcas] |
| 5 |
Hb_000223_050 |
0.13468942 |
- |
- |
PREDICTED: endochitinase A-like [Jatropha curcas] |
| 6 |
Hb_001838_010 |
0.1352492075 |
- |
- |
hypothetical protein POPTR_0005s18380g [Populus trichocarpa] |
| 7 |
Hb_000723_120 |
0.1360889886 |
- |
- |
PREDICTED: UDP-glycosyltransferase 86A1 [Jatropha curcas] |
| 8 |
Hb_008948_080 |
0.141322261 |
- |
- |
PREDICTED: TBC1 domain family member 13-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_000625_060 |
0.1423461944 |
- |
- |
Protein P21, putative [Ricinus communis] |
| 10 |
Hb_011214_140 |
0.142610162 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 11 |
Hb_000120_070 |
0.1442995501 |
- |
- |
PREDICTED: uncharacterized protein LOC105630351 [Jatropha curcas] |
| 12 |
Hb_002917_030 |
0.1457136226 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase HSL2 [Setaria italica] |
| 13 |
Hb_001355_110 |
0.1468201927 |
- |
- |
PREDICTED: mitochondrial chaperone BCS1-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_000489_020 |
0.1486575415 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Populus euphratica] |
| 15 |
Hb_012753_200 |
0.1497547432 |
- |
- |
hypothetical protein JCGZ_20651 [Jatropha curcas] |
| 16 |
Hb_000579_180 |
0.1498062825 |
transcription factor |
TF Family: bZIP |
PREDICTED: ocs element-binding factor 1-like [Jatropha curcas] |
| 17 |
Hb_002805_150 |
0.1498191579 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 18 |
Hb_000512_080 |
0.1499699288 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 19 |
Hb_000836_390 |
0.153354247 |
- |
- |
PREDICTED: neurofilament light polypeptide-like [Gossypium raimondii] |
| 20 |
Hb_000676_030 |
0.1535893527 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-B [Jatropha curcas] |